The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis
- PMID: 31586073
- PMCID: PMC6778137
- DOI: 10.1038/s41467-019-12507-9
The midbody interactome reveals unexpected roles for PP1 phosphatases in cytokinesis
Abstract
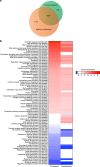
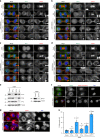
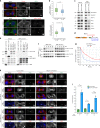
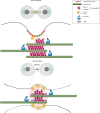
The midbody is an organelle assembled at the intercellular bridge between the two daughter cells at the end of mitosis. It controls the final separation of the daughter cells and has been involved in cell fate, polarity, tissue organization, and cilium and lumen formation. Here, we report the characterization of the intricate midbody protein-protein interaction network (interactome), which identifies many previously unknown interactions and provides an extremely valuable resource for dissecting the multiple roles of the midbody. Initial analysis of this interactome revealed that PP1β-MYPT1 phosphatase regulates microtubule dynamics in late cytokinesis and de-phosphorylates the kinesin component MKLP1/KIF23 of the centralspindlin complex. This de-phosphorylation antagonizes Aurora B kinase to modify the functions and interactions of centralspindlin in late cytokinesis. Our findings expand the repertoire of PP1 functions during mitosis and indicate that spatiotemporal changes in the distribution of kinases and counteracting phosphatases finely tune the activity of cytokinesis proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

