Transitioning to composite bacterial mutagenicity models in ICH M7 (Q)SAR analyses
- PMID: 31586682
- PMCID: PMC6919322
- DOI: 10.1016/j.yrtph.2019.104488
Transitioning to composite bacterial mutagenicity models in ICH M7 (Q)SAR analyses
Abstract
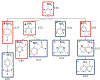
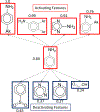
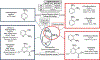
The International Council on Harmonisation (ICH) M7(R1) guideline describes the use of complementary (quantitative) structure-activity relationship ((Q)SAR) models to assess the mutagenic potential of drug impurities in new and generic drugs. Historically, the CASE Ultra and Leadscope software platforms used two different statistical-based models to predict mutations at G-C (guanine-cytosine) and A-T (adenine-thymine) sites, to comprehensively assess bacterial mutagenesis. In the present study, composite bacterial mutagenicity models covering multiple mutation types were developed. These new models contain more than double the number of chemicals (n = 9,254 and n = 13,514) than the corresponding non-composite models and show better toxicophore coverage. Additionally, the use of a single composite bacterial mutagenicity model simplifies impurity analysis in an ICH M7 (Q)SAR workflow by reducing the number of model outputs requiring review. An external validation set of 388 drug impurities representing proprietary pharmaceutical chemical space showed performance statistics ranging from of 66-82% in sensitivity, 91-95% in negative predictivity and 96% in coverage. This effort represents a major enhancement to these (Q)SAR models and their use under ICH M7(R1), leading to improved patient safety through greater predictive accuracy, applicability, and efficiency when assessing the bacterial mutagenic potential of drug impurities.
Keywords: Ames; Bacterial mutagenicity; Computational toxicology; Drug; Genotoxicity; ICH M7; In vitro; QSAR; Regulatory review; Structure-activity relationship.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
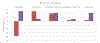

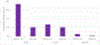

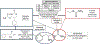

References
-
- Ahlberg E; Amberg A; Beilke LD; Bower D; Cross KP; Custer L; Ford KA; Van Gompel J; Harvey J; Honma M; Jolly R; Joossens E; Kemper RA; Kenyon M; Kruhlak N; Kuhnke L; Leavitt P; Naven R; Neilan C; Quigley DP; Shuey D; Spirkl HP; Stavitskaya L; Teasdale A; White A; Wichard J; Zwickl C; Myatt GJ, Extending (Q)SARs to incorporate proprietary knowledge for regulatory purposes: A case study using aromatic amine mutagenicity. Regul Toxicol Pharmacol 2016, 77, 1–12. https://doi.org/10.1016Z.yrtph.2016.02.003 - PubMed
-
- Amberg A; Andaya RV; Anger LT; Barber C; Beilke L; Bercu J; Bower D; Brigo A; Cammerer Z; Cross KP; Custer L; Dobo K; Gerets H; Gervais V; Glowienke S; Gomez S; Van Gompel J; Harvey J; Hasselgren C; Honma M; Johnson C; Jolly R; Kemper R; Kenyon M; Kruhlak N; Leavitt P; Miller S; Muster W; Naven R; Nicolette J; Parenty A; Powley M; Quigley DP; Reddy MV; Sasaki JC; Stavitskaya L; Teasdale A; Trejo-Martin A; Weiner S; Welch DS; White A; Wichard J; Woolley D; Myatt GJ, Principles and procedures for handling out-of-domain and indeterminate results as part of ICH M7 recommended (Q)SAR analyses. Regul Toxicol Pharmacol 2019, 102, 53–64. https://doi.org/10.1016Z.yrtph.2018.12.007 - PMC - PubMed
-
- Amberg A; Beilke L; Bercu J; Bower D; Brigo A; Cross KP; Custer L; Dobo K; Dowdy E; Ford KA; Glowienke S; Van Gompel J; Harvey J; Hasselgren C; Honma M; Jolly R; Kemper R; Kenyon M; Kruhlak N; Leavitt P; Miller S; Muster W; Nicolette J; Plaper A; Powley M; Quigley DP; Reddy MV; Spirkl HP; Stavitskaya L; Teasdale A; Weiner S; Welch DS; White A; Wichard J; Myatt GJ, Principles and procedures for implementation of ICH M7 recommended (Q)SAR analyses. Regul Toxicol Pharmacol 2016, 77, 13–24. 10.1016/jlyrtph.2016.02.004 - DOI - PubMed
-
- Amberg A; Harvey JS; Czich A; Spirkl H-P; Robinson S; White A; Elder DP, Do Carboxylic/Sulfonic Acid Halides Really Present a Mutagenic and Carcinogenic Risk as Impurities in Final Drug Products? Organic Process Research & Development 2015, 19 (11), 1495–1506. 10.1021/acs.oprd.5b00106 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

