Alternative splicing of VEGFA is regulated by RBM10 in endometrial cancer
- PMID: 31587503
- PMCID: PMC11896373
- DOI: 10.1002/kjm2.12127
Alternative splicing of VEGFA is regulated by RBM10 in endometrial cancer
Abstract
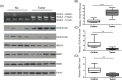
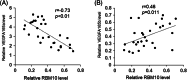
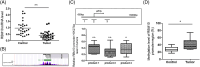
Vascular endothelial growth factor A (VEGFA) gene has three alternative exons which results in multiple isoforms. VEGFA has been found overexpressed in patients with endometrial cancer, but the VEGFA expression pattern and how it is regulated are still unknown. The level of VEGFA transcripts and protein isoforms were detected by semi-quantitative Polymerase chain reaction (PCR) and immunoblotting in 29 paired endometrial tumor and adjacent nontumor control tissues. The level of three alternative splicing related proteins: RBM5, RBM6, and RBM10 was determined by immunoblotting. The H3K27Ac level in RBM10 promoter region was detected by ChIP-PCR. The RBM10 promoter region methylation level were quantified by methylation-sensitive high resolution melting. VEGFA165a was overexpressed and VEGFA165b level was reduced in tumors. RBM10 level was reduced in tumors. RBM10 level was negatively correlated with VEGFA165a level and positively correlated with VEGFA165b level in tumors. Using HEC-1-A and RL95-2 cells, we confirmed that VEGFA165a/b expressed pattern was controlled by RBM10. MALAT1 level was increased in tumors but not involved in VEGFA alternative splicing. Reduced H3K27Ac level and increased DNA methylation in the promoter region controlled RBM10 expression in tumors. VEGFA alternative splicing in endometrial cancer was regulated by RBM10, the expression of which was controlled by histone acetylation and DNA methylation.
Keywords: MALAT1; RBM10; VEGFA; alternative splicing; endometrial cancer.
© 2019 The Authors. The Kaohsiung Journal of Medical Sciences published by John Wiley & Sons Australia on behalf of Kaohsiung Medical University.
Conflict of interest statement
All authors declare no conflict of interest.
Figures

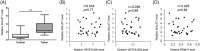
Similar articles
-
RBM10: Structure, functions, and associated diseases.Gene. 2021 May 30;783:145463. doi: 10.1016/j.gene.2021.145463. Epub 2021 Jan 28. Gene. 2021. PMID: 33515724 Free PMC article. Review.
-
miR-335 modulates Numb alternative splicing via targeting RBM10 in endometrial cancer.Kaohsiung J Med Sci. 2020 Mar;36(3):171-177. doi: 10.1002/kjm2.12149. Epub 2020 Jan 2. Kaohsiung J Med Sci. 2020. PMID: 31894898 Free PMC article.
-
Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay.Nucleic Acids Res. 2017 Aug 21;45(14):8524-8540. doi: 10.1093/nar/gkx508. Nucleic Acids Res. 2017. PMID: 28586478 Free PMC article.
-
Expression of RBM5-related factors in primary breast tissue.J Cell Biochem. 2007 Apr 15;100(6):1440-58. doi: 10.1002/jcb.21134. J Cell Biochem. 2007. PMID: 17131366
-
RBM5 as a putative tumor suppressor gene for lung cancer.J Thorac Oncol. 2010 Mar;5(3):294-8. doi: 10.1097/JTO.0b013e3181c6e330. J Thorac Oncol. 2010. PMID: 20186023 Review.
Cited by
-
RBM10: Structure, functions, and associated diseases.Gene. 2021 May 30;783:145463. doi: 10.1016/j.gene.2021.145463. Epub 2021 Jan 28. Gene. 2021. PMID: 33515724 Free PMC article. Review.
-
Genome-Wide Identification of Immune-Related Alternative Splicing and Splicing Regulators Involved in Abdominal Aortic Aneurysm.Front Genet. 2022 Feb 17;13:816035. doi: 10.3389/fgene.2022.816035. eCollection 2022. Front Genet. 2022. PMID: 35251127 Free PMC article.
-
Alternative splicing and cancer: a systematic review.Signal Transduct Target Ther. 2021 Feb 24;6(1):78. doi: 10.1038/s41392-021-00486-7. Signal Transduct Target Ther. 2021. PMID: 33623018 Free PMC article.
-
Alternative splicing in endothelial cells: novel therapeutic opportunities in cancer angiogenesis.J Exp Clin Cancer Res. 2020 Dec 7;39(1):275. doi: 10.1186/s13046-020-01753-1. J Exp Clin Cancer Res. 2020. PMID: 33287867 Free PMC article. Review.
-
Harnessing DNA replication stress to target RBM10 deficiency in lung adenocarcinoma.Nat Commun. 2024 Jul 30;15(1):6417. doi: 10.1038/s41467-024-50882-0. Nat Commun. 2024. PMID: 39080280 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. - PubMed
-
- Saarelainen SK, Staff S, Peltonen N, Lehtimaki T, Isola J, Kujala PM, et al. Endoglin, VEGF, and its receptors in predicting metastases in endometrial carcinoma. Tumour Biol. 2014;35(5):4651–4657. - PubMed
-
- Topolovec Z, Corusic A, Babic D, Mrcela M, Sijanovic S, Muller‐Vranjes A, et al. Vascular endothelial growth factor and intratumoral microvessel density as prognostic factors in endometrial cancer. Coll Antropol. 2010;34(2):447–453. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

