Full-Length P2X7 Structures Reveal How Palmitoylation Prevents Channel Desensitization
- PMID: 31587896
- PMCID: PMC7053488
- DOI: 10.1016/j.cell.2019.09.017
Full-Length P2X7 Structures Reveal How Palmitoylation Prevents Channel Desensitization
Abstract
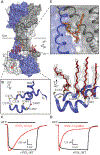
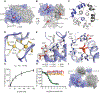
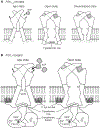
P2X receptors are trimeric, non-selective cation channels activated by extracellular ATP. The P2X7 receptor subtype is a pharmacological target because of involvement in apoptotic, inflammatory, and tumor progression pathways. It is the most structurally and functionally distinct P2X subtype, containing a unique cytoplasmic domain critical for the receptor to initiate apoptosis and not undergo desensitization. However, lack of structural information about the cytoplasmic domain has hindered understanding of the molecular mechanisms underlying these processes. We report cryoelectron microscopy structures of full-length rat P2X7 receptor in apo and ATP-bound states. These structures reveal how one cytoplasmic element, the C-cys anchor, prevents desensitization by anchoring the pore-lining helix to the membrane with palmitoyl groups. They show a second cytoplasmic element with a unique fold, the cytoplasmic ballast, which unexpectedly contains a zinc ion complex and a guanosine nucleotide binding site. Our structures provide first insights into the architecture and function of a P2X receptor cytoplasmic domain.
Keywords: Apoptosis; Cryoelectron microscopy; Cytoplasmic domain; Desensitization; Guanosine nucleotides; Ligand gated ion channels; Palmitoylation; Purinergic (P2X) receptors; Structural biology; Zinc ion complex.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures

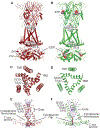


References
-
- Adinolfi E, Cirillo M, Woltersdorf R, Falzoni S, Chiozzi P, Pellegatti P, Callegari MG, Sandona D, Markwardt F, Schmalzing G, et al. (2010). Trophic activity of a naturally occurring truncated isoform of the P2X7 receptor. FASEB J 24, 3393–3404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

