Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity
- PMID: 31588020
- PMCID: PMC6944069
- DOI: 10.1016/j.ccell.2019.09.001
Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity
Erratum in
-
Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity.Cancer Cell. 2020 Mar 16;37(3):420. doi: 10.1016/j.ccell.2020.03.003. Cancer Cell. 2020. PMID: 32183953 Free PMC article. No abstract available.
Abstract
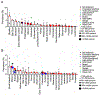
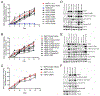
We characterized the landscape and drug sensitivity of ERBB2 (HER2) mutations in cancers. In 11 datasets (n = 211,726), ERBB2 mutational hotspots varied across 25 tumor types. Common HER2 mutants yielded differential sensitivities to eleven EGFR/HER2 tyrosine kinase inhibitors (TKIs) in vitro, and molecular dynamics simulations revealed that mutants with a reduced drug-binding pocket volume were associated with decreased affinity for larger TKIs. Overall, poziotinib was the most potent HER2 mutant-selective TKI tested. Phase II clinical testing in ERBB2 exon 20-mutant non-small cell lung cancer resulted in a confirmed objective response rate of 42% in the first 12 evaluable patients. In pre-clinical models, poziotinib upregulated HER2 cell-surface expression and potentiated the activity of T-DM1, resulting in complete tumor regression with combination treatment.
Keywords: ERBB2 mutant; HER2 mutant; NSCLC; T-DM1; TKI; exon 20; pan-cancer; poziotinib.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests
The research being reported in this publication is research in which The University of Texas MD Anderson Cancer Center has an institutional financial conflict of interest. Because MD Anderson is committed to the protection of human subjects and the effective management of its financial conflicts of interest in relation to its research activities, MD Anderson has implemented an Institutional Conflict of Interest Management and Monitoring Plan to manage and monitor the conflict of interest with respect to MD Anderson’ s conduct of this research.
Figures






References
-
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, et al. (2010). Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376, 687–697. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

