Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer
- PMID: 31588021
- PMCID: PMC6801104
- DOI: 10.1016/j.ccell.2019.08.007
Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer
Abstract
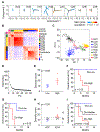
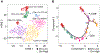
Cellular diversity in tumors is a key factor for therapeutic failures and lethal outcomes of solid malignancies. Here, we determined the single-cell transcriptomic landscape of liver cancer biospecimens from 19 patients. We found varying degrees of heterogeneity in malignant cells within and between tumors and diverse landscapes of tumor microenvironment (TME). Strikingly, tumors with higher transcriptomic diversity were associated with patient's worse overall survival. We found a link between hypoxia-dependent vascular endothelial growth factor expression in tumor diversity and TME polarization. Moreover, T cells from higher heterogeneous tumors showed lower cytolytic activities. Consistent results were found using bulk genomic and transcriptomic profiles of 765 liver tumors. Our results offer insight into the diverse ecosystem of liver cancer and its impact on patient prognosis.
Keywords: VEGF; biodiversity; cholangiocarcinoma; hepatocellular carcinoma; liver cancer; microenvironmental reprogramming; single-cell; tumor ecosystem; tumor heterogeneity; tumor microenvironments.
Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




References
-
- Baslan T, and Hicks J (2017). Unravelling biology and shifting paradigms in cancer with single-cell sequencing. Nat Rev Cancer 17, 557–569. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

