The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis
- PMID: 31588046
- PMCID: PMC7247364
- DOI: 10.1016/j.stem.2019.08.018
The RNA Helicase DDX6 Controls Cellular Plasticity by Modulating P-Body Homeostasis
Abstract
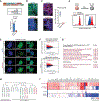
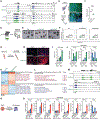
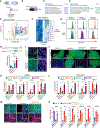
Post-transcriptional mechanisms have the potential to influence complex changes in gene expression, yet their role in cell fate transitions remains largely unexplored. Here, we show that suppression of the RNA helicase DDX6 endows human and mouse primed embryonic stem cells (ESCs) with a differentiation-resistant, "hyper-pluripotent" state, which readily reprograms to a naive state resembling the preimplantation embryo. We further demonstrate that DDX6 plays a key role in adult progenitors where it controls the balance between self-renewal and differentiation in a context-dependent manner. Mechanistically, DDX6 mediates the translational suppression of target mRNAs in P-bodies. Upon loss of DDX6 activity, P-bodies dissolve and release mRNAs encoding fate-instructive transcription and chromatin factors that re-enter the ribosome pool. Increased translation of these targets impacts cell fate by rewiring the enhancer, heterochromatin, and DNA methylation landscapes of undifferentiated cell types. Collectively, our data establish a link between P-body homeostasis, chromatin organization, and stem cell potency.
Keywords: P-body; RNA helicase DDX6; adult progenitor cells; chromatin; differentiation; embryonic stem cells; exit from pluripotency; naive pluripotency; post-transcriptional regulation; primed pluripotency; self-renewal.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




Comment in
-
The Princess and the P: Pluripotent Stem Cells and P-Bodies.Cell Stem Cell. 2019 Nov 7;25(5):589-591. doi: 10.1016/j.stem.2019.10.008. Cell Stem Cell. 2019. PMID: 31703768
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

