VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium
- PMID: 31588507
- PMCID: PMC6943061
- DOI: 10.1093/nar/gkz874
VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium
Abstract
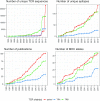
Here, we report an update of the VDJdb database with a substantial increase in the number of T-cell receptor (TCR) sequences and their cognate antigens. The update further provides a new database infrastructure featuring two additional analysis modes that facilitate database querying and real-world data analysis. The increased yield of TCR specificity identification methods and the overall increase in the number of studies in the field has allowed us to expand the database more than 5-fold. Furthermore, several new analysis methods are included. For example, batch annotation of TCR repertoire sequencing samples allows for annotating large datasets on-line. Using recently developed bioinformatic methods for TCR motif mining, we have built a reduced set of high-quality TCR motifs that can be used for both training TCR specificity predictors and matching against TCRs of interest. These additions enhance the versatility of the VDJdb in the task of exploring T-cell antigen specificities. The database is available at https://vdjdb.cdr3.net.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Rojas M., Restrepo-Jiménez P., Monsalve D.M., Pacheco Y., Acosta-Ampudia Y., Ramírez-Santana C., Leung P.S.C., Ansari A.A., Gershwin M.E., Anaya J.-M.. Molecular mimicry and autoimmunity. J. Autoimmun. 2018; 95:100–123. - PubMed
-
- Bentzen A.K., Marquard A.M., Lyngaa R., Saini S.K., Ramskov S., Donia M., Such L., Furness A.J.S., McGranahan N., Rosenthal R. et al. .. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 2016; 34:1037–1045. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

