Proximal tubule ATR regulates DNA repair to prevent maladaptive renal injury responses
- PMID: 31589169
- PMCID: PMC6819104
- DOI: 10.1172/JCI122313
Proximal tubule ATR regulates DNA repair to prevent maladaptive renal injury responses
Abstract
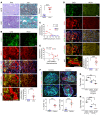
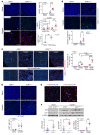
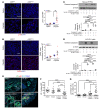
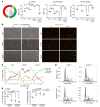
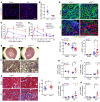
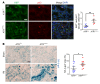
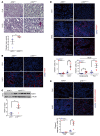
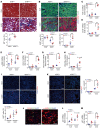
Maladaptive proximal tubule (PT) repair has been implicated in kidney fibrosis through induction of cell-cycle arrest at G2/M. We explored the relative importance of the PT DNA damage response (DDR) in kidney fibrosis by genetically inactivating ataxia telangiectasia and Rad3-related (ATR), which is a sensor and upstream initiator of the DDR. In human chronic kidney disease, ATR expression inversely correlates with DNA damage. ATR was upregulated in approximately 70% of Lotus tetragonolobus lectin-positive (LTL+) PT cells in cisplatin-exposed human kidney organoids. Inhibition of ATR resulted in greater PT cell injury in organoids and cultured PT cells. PT-specific Atr-knockout (ATRRPTC-/-) mice exhibited greater kidney function impairment, DNA damage, and fibrosis than did WT mice in response to kidney injury induced by either cisplatin, bilateral ischemia-reperfusion, or unilateral ureteral obstruction. ATRRPTC-/- mice had more cells in the G2/M phase after injury than did WT mice after similar treatments. In conclusion, PT ATR activation is a key component of the DDR, which confers a protective effect mitigating the maladaptive repair and consequent fibrosis that follow kidney injury.
Keywords: Cell cycle; Chronic kidney disease; Fibrosis; Nephrology.
Conflict of interest statement
Figures

Comment in
-
DNA damage response protects against progressive kidney disease.J Clin Invest. 2019 Nov 1;129(11):4574-4575. doi: 10.1172/JCI131171. J Clin Invest. 2019. PMID: 31589167 Free PMC article.
References
-
- [No authors listed]. Chronic kidney disease fact sheet. World Kidney Day. https://www.worldkidneyday.org/facts/chronic-kidney-disease/ Accessed August 14, 2019.
Publication types
MeSH terms
Substances
Grants and funding
- K01 DK099473/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R37 DK039773/DK/NIDDK NIH HHS/United States
- R01 DK072381/DK/NIDDK NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- UG3 TR002155/TR/NCATS NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- R01 DK121101/DK/NIDDK NIH HHS/United States
- UH3 TR002155/TR/NCATS NIH HHS/United States
- T32 DK007527/DK/NIDDK NIH HHS/United States
- P30 DK114809/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

