Pyrin dephosphorylation is sufficient to trigger inflammasome activation in familial Mediterranean fever patients
- PMID: 31589380
- PMCID: PMC6835204
- DOI: 10.15252/emmm.201910547
Pyrin dephosphorylation is sufficient to trigger inflammasome activation in familial Mediterranean fever patients
Abstract
Familial Mediterranean fever (FMF) is the most frequent hereditary systemic autoinflammatory syndrome. FMF is usually caused by biallelic mutations in the MEFV gene, encoding Pyrin. Conclusive genetic evidence lacks for about 30% of patients diagnosed with clinical FMF. Pyrin is an inflammasome sensor maintained inactive by two kinases (PKN1/2). The consequences of MEFV mutations on inflammasome activation are still poorly understood. Here, we demonstrate that PKC superfamily inhibitors trigger inflammasome activation in monocytes from FMF patients while they trigger a delayed apoptosis in monocytes from healthy donors. The expression of the pathogenic p.M694V MEFV allele is necessary and sufficient for PKC inhibitors (or mutations precluding Pyrin phosphorylation) to trigger caspase-1- and gasdermin D-mediated pyroptosis. In line with colchicine efficacy in patients, colchicine fully blocks this response in FMF patients' monocytes. These results indicate that Pyrin inflammasome activation is solely controlled by Pyrin (de)phosphorylation in FMF patients while a second control mechanism restricts its activation in healthy donors/non-FMF patients. This study paves the way toward a functional characterization of MEFV variants and a functional test to diagnose FMF.
Keywords: autoinflammation; caspase-1; colchicine; diagnosis; pyroptosis.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
FM, LL, AM, AB, YJ, and TH are listed on a patent related to FMF diagnosis.
Figures

- A–I
Monocytes from healthy donors (HD) or FMF patients were either primed with LPS (A–C, G–I) or not (D–F) and treated with (A, C) 1.25 μM staurosporine (Stauro), (B, D–F) 12.5 μM UCN‐01, or (G–I) 5 μM nigericin (Nig). (A, B) IL‐1β and (C) TNF level were quantified by ELISA at 90 min post stimulation. (D, G) Cell death was monitored in real time by measuring propidium iodide influx/fluorescence every 5 min. (E, H) The time required to reach 20% cell death and (F, I) the area under the curve (AUC) were computed for each HD or FMF patients.
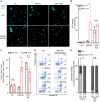
- A, B
Monocytes from HD or FMF patients were treated with 12.5 μM UCN‐01 for 40 min or primed with LPS (3 h) and treated with 5 μM nigericin (Nig) for 90 min. (A) Cells were immuno‐stained for ASC. ASC specks are indicated by red arrowheads. Representative confocal microscopy images from one HD (top panels) and one FMF patient (bottom panels) are shown. Scale bars: 10 μm and 2.5 μm in the main figures and onsets, respectively. (B) Quantification of ASC specks in HD and FMF patients’ monocytes by immunofluorescence.
- C
The frequency of cells positive for active caspase‐1 was quantified by flow cytometry, using FLICA‐Caspase‐1 in HD and FMF patients.
- D, E
Cell death was assessed at 90 min post‐UCN‐01 or post‐LPS + nigericin treatment by determining the percentage of Annexin‐V+/PI− cells and of PI+ cells among dead cells (Annexin‐V+ and/or PI + cells) using flow cytometry. (D) Representative FACS plots from one healthy donor (HD) and one FMF patient are shown. Percentage are indicated for the two right gates. (E) Cell death modality was assessed by determining the percentage of treatment‐induced Annexin‐V+/PI− cells and of PI+ cells among dead cells, using flow cytometry.

- A, B
IL‐1β level was quantified by ELISA at 90 min post‐stimulation.
- C–E
Cell death was monitored in real time by measuring propidium iodide influx/fluorescence every 5 min.
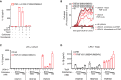
- A
Primary human monocytes from FMF patient were primed with LPS and stimulated as indicated with UCN‐01 in the presence of paclitaxel (Taxol, 5 μM), nocodazole (5 μM), or colchicine (1 μM).
- B
Propidium iodide incorporation was monitored every 5 min post‐UCN‐01 addition in the presence of Taxol (5 μM), nocodazole (5 μM), or colchicine (1 μM). PI incorporation was normalized using TX‐100 cells (total PI incorporation). (A, C, D) IL‐1β concentration in the supernatant was quantified by ELISA.
- C, D
Primary human monocytes from the indicated healthy donor (HD) or FMF patient were primed with LPS and stimulated as indicated with (C) UCN‐01 or (D) TcdA (1 μg/ml) in the presence of the indicated concentration of colchicine.

- A, B
The expression of Flag‐Pyrin was revealed by Western blotting analysis against Flag (A) or against Pyrin (B).
- C
IL‐18 levels were quantified in the supernatant of PMA‐differentiated U937 cells by ELISA at 3 h post‐treatment with UCN‐01 or staurosporine (Stauro). One experiment representative of two independent experiments is shown. The bar represents the mean of a biological triplicate.

- A–C
U937 cells of the indicated genotype and expressing the indicated plasmids were treated with (A, C) UCN‐01 or LPS + nigericin (Nig). Propidium iodide (PI) influx/fluorescence was monitored every 5 min for 3 h.
- D, E
PMA‐differentiated U937 cells of the indicated genotype and expressing the indicated plasmids were primed with LPS for 3 h and treated with UCN‐01, staurosporine (Stauro) or nigericin as indicated. IL‐1β level in the supernatant was quantified by ELISA at 3 h post‐treatment.
- F
Pyrin S242 phosphorylation was assessed by Western blot in the indicated cell lines with and without UCN‐01 treatment for 15 min.
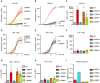
- A–F
Propidium iodide (PI) influx/fluorescence was monitored every 5 min for 3 h in response to UCN‐01 (A–C) or LPS + nigericin (D–F).
- G, H
U937 cells were differentiated in macrophages using PMA, treated with LPS and UCN‐01 (G) or nigericin (H). IL‐1β levels were quantified by ELISA.
- I
The frequency of the different MEFV alleles is shown (data extracted from gnomad).
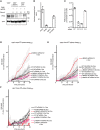
- A
PKN1KO clones expressing p.M694V or WT Pyrin were obtained and confirmed by Western blot analysis. Four clones are shown. Clones 3 and 6 were selected.
- B
Absolute transcript level of the indicated genes was quantified by RNAseq in U937 cells (Benaoudia et al, 2019) and expressed as count per million (cpm). PYCARD encodes ASC and is shown as a reference.
- C
PKN2 transcript levels were assessed in U937 cells at 24 h post‐electroporation with the indicated non‐targeting (NT) siRNA or three different siRNAs targeting PKN2.
- D–F
U937 cells with the indicated Pyrin variant were treated (+Dox, plain lines) or not (No Dox, dotted lines) with doxycycline (Dox) at 24 h post‐electroporation with the indicated siRNA. Cell death was monitored every 15 min. (F) SiRNA 14 did not reduce substantially PKN2 levels.

- A, B
Propidium iodide (PI) influx/fluorescence was monitored every 15 min for 20 h and every 5 min for an additional 4 h.
- C, D
Area under the curve (AUC) corresponding to Fig 5A and B was computed from t = 0 to t = 20 h (C) and from 20 to 24 h (D).
- E, F
IL‐18 was quantified in the supernatant of the indicated U937 cells at 15 h post‐doxycycline/PBS addition (E) or in the supernatant of U937 cells exposed to 15 h of doxycycline/PBS and an additional 3 h of incubation with UCN‐01 (F).

- A
Propidium iodide (PI) influx/fluorescence was monitored every 15 min for 20 h.
- B
Area under the curve (AUC) corresponding to Fig EV5A is shown.

- A, B
IL‐1β level was quantified by ELISA at 90 min post‐stimulation.
- C–E
Cell death was monitored in real time by measuring propidium iodide (PI) influx/fluorescence every 5 min. (D) The time required to obtain 20% of cell death and (E) the area under the kinetics curve (AUC) were computed for each patient.
- F–I
Receiver operating characteristic (ROC) curves were computed for IL‐1β concentrations following (F) staurosporine or (G) UCN‐01 treatment, (H) the time to obtain 20% cell death, and (I) the area under the cell death kinetics curve. For each ROC curve, the AUC, the positive (ppv), and negative (npv) predictive values are indicated.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

