Model-based physiological noise removal in fast fMRI
- PMID: 31589991
- PMCID: PMC6911832
- DOI: 10.1016/j.neuroimage.2019.116231
Model-based physiological noise removal in fast fMRI
Abstract
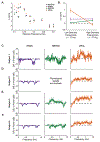
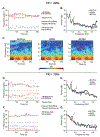
Recent improvements in the speed and sensitivity of fMRI acquisition techniques suggest that fast fMRI can be used to detect and precisely localize sub-second neural dynamics. This enhanced temporal resolution has enormous potential for neuroscientists. However, physiological noise poses a major challenge for the analysis of fast fMRI data. Physiological noise scales with sensitivity, and its autocorrelation structure is altered in rapidly sampled data, suggesting that new approaches are needed for physiological noise removal in fast fMRI. Existing strategies either rely on external physiological recordings, which can be noisy or difficult to collect, or employ data-driven approaches which make assumptions that may not hold true in fast fMRI. We created a statistical model of harmonic regression with autoregressive noise (HRAN) to estimate and remove cardiac and respiratory noise from the fMRI signal directly. This technique exploits the fact that cardiac and respiratory noise signals are fully sampled (rather than aliasing) when imaging at fast rates, allowing us to track and model physiology over time without requiring external physiological measurements. We then created a joint model of neural hemodynamics, and physiological and autocorrelated noise to more accurately remove noise. We first verified that HRAN accurately estimates cardiac and respiratory dynamics and that our model demonstrates goodness-of-fit in fast fMRI data. In task-driven data, we then demonstrated that HRAN is able to remove physiological noise while leaving the neural signal intact, thereby increasing detection of task-driven voxels. Finally, we established that in both simulations and fast fMRI data HRAN is able to improve statistical inferences as compared with gold-standard physiological noise removal techniques. In conclusion, we created a tool that harnesses the novel information in fast fMRI to remove physiological noise, enabling broader use of the technology to study human brain function.
Keywords: Autocorrelation; Fast fMRI; HRAN; Harmonic regression; Physiological noise; Simultaneous multislice (SMS).
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of Interest
The authors have no competing interests to declare.
Figures


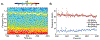




References
-
- Aslan Serdar, Hocke Lia, Schwarz Nicolette, and Frederick Blaise. “Extraction of the Cardiac Waveform from Simultaneous Multislice FMRI Data Using Slice Sorted Averaging and a Deep Learning Reconstruction Filter.” NeuroImage 198 (September 1, 2019): 303–16. 10.1016/j.neuroimage.2019.05.049. - DOI - PMC - PubMed
-
- Barbieri Riccardo, Matten Eric C., Alabi AbdulRasheed A., and Brown Emery N.. “A Point-Process Model of Human Heartbeat Intervals: New Definitions of Heart Rate and Heart Rate Variability.” American Journal of Physiology-Heart and Circulatory Physiology 288, no. 1 (January 2004): H424–35. 10.1152/ajpheart.00482.2003. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

