Bringing radiomics into a multi-omics framework for a comprehensive genotype-phenotype characterization of oncological diseases
- PMID: 31590671
- PMCID: PMC6778975
- DOI: 10.1186/s12967-019-2073-2
Bringing radiomics into a multi-omics framework for a comprehensive genotype-phenotype characterization of oncological diseases
Abstract
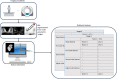
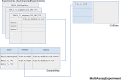
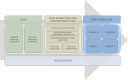
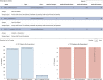
Genomic and radiomic data integration, namely radiogenomics, can provide meaningful knowledge in cancer diagnosis, prognosis and treatment. Despite several data structures based on multi-layer architecture proposed to combine multi-omic biological information, none of these has been designed and assessed to include radiomic data as well. To meet this need, we propose to use the MultiAssayExperiment (MAE), an R package that provides data structures and methods for manipulating and integrating multi-assay experiments, as a suitable tool to manage radiogenomic experiment data. To this aim, we first examine the role of radiogenomics in cancer phenotype definition, then the current state of radiogenomics data integration in public repository and, finally, challenges and limitations of including radiomics in MAE, designing an extended framework and showing its application on a case study from the TCGA-TCIA archives. Radiomic and genomic data from 91 patients have been successfully integrated in a single MAE object, demonstrating the suitability of the MAE data structure as container of radiogenomic data.
Keywords: Cancer; MultiAssayExperiment; Radiogenomics; Radiomics; TCGA; TCIA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

