A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation
- PMID: 31591036
- PMCID: PMC6906606
- DOI: 10.1016/j.chembiol.2019.09.010
A Catalytic Trisulfide in Human Sulfide Quinone Oxidoreductase Catalyzes Coenzyme A Persulfide Synthesis and Inhibits Butyrate Oxidation
Abstract
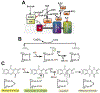
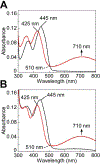
Mitochondrial sulfide quinone oxidoreductase (SQR) catalyzes the oxidation of H2S to glutathione persulfide with concomitant reduction of CoQ10. We report herein that the promiscuous activity of human SQR supported the conversion of CoA to CoA-SSH (CoA-persulfide), a potent inhibitor of butyryl-CoA dehydrogenase, and revealed a molecular link between sulfide and butyrate metabolism, which are known to interact. Three different CoQ1-bound crystal structures furnished insights into how diverse substrates access human SQR, and provided snapshots of the reaction coordinate. Unexpectedly, the active site cysteines in SQR are configured in a bridging trisulfide at the start and end of the catalytic cycle, and the presence of sulfane sulfur was confirmed biochemically. Importantly, our study leads to a mechanistic proposal for human SQR in which sulfide addition to the trisulfide cofactor eliminates 201Cys-SSH, forming an intense charge-transfer complex with flavin adenine dinucleotide, and 379Cys-SSH, which transfers sulfur to an external acceptor.
Keywords: butyrate; coenzyme Q; crystal structure; enzyme kinetics; flavin; hydrogen sulfide.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interest
The authors declare that no competing financial interests exist.
Figures




Similar articles
-
Staphylococcus aureus sqr Encodes a Type II Sulfide:Quinone Oxidoreductase and Impacts Reactive Sulfur Speciation in Cells.Biochemistry. 2016 Nov 29;55(47):6524-6534. doi: 10.1021/acs.biochem.6b00714. Epub 2016 Nov 16. Biochemistry. 2016. PMID: 27806570 Free PMC article.
-
H2S oxidation by nanodisc-embedded human sulfide quinone oxidoreductase.J Biol Chem. 2017 Jul 14;292(28):11641-11649. doi: 10.1074/jbc.M117.788547. Epub 2017 May 16. J Biol Chem. 2017. PMID: 28512131 Free PMC article.
-
Transient Kinetic Analysis of Hydrogen Sulfide Oxidation Catalyzed by Human Sulfide Quinone Oxidoreductase.J Biol Chem. 2015 Oct 9;290(41):25072-80. doi: 10.1074/jbc.M115.682369. Epub 2015 Aug 28. J Biol Chem. 2015. PMID: 26318450 Free PMC article.
-
Hydrogen Sulfide Oxidation by Sulfide Quinone Oxidoreductase.Chembiochem. 2021 Mar 16;22(6):949-960. doi: 10.1002/cbic.202000661. Epub 2020 Nov 17. Chembiochem. 2021. PMID: 33080111 Free PMC article. Review.
-
A new structure-based classification of sulfide:quinone oxidoreductases.Proteins. 2010 Apr;78(5):1073-83. doi: 10.1002/prot.22665. Proteins. 2010. PMID: 20077566 Review.
Cited by
-
Mitochondrial Dysfunction and Redox Homeostasis Impairment as Pathomechanisms of Brain Damage in Ethylmalonic Encephalopathy: Insights from Animal and Human Studies.Cell Mol Neurobiol. 2022 Apr;42(3):565-575. doi: 10.1007/s10571-020-00976-2. Epub 2020 Oct 9. Cell Mol Neurobiol. 2022. PMID: 33034777 Free PMC article. Review.
-
Regulation of redox signaling by reactive sulfur species.J Clin Biochem Nutr. 2021 Mar;68(2):111-115. doi: 10.3164/jcbn.20-124. Epub 2021 Feb 17. J Clin Biochem Nutr. 2021. PMID: 33879961 Free PMC article. Japanese.
-
Gas regulation of complex II reversal via electron shunting to fumarate in the mammalian ETC.Trends Biochem Sci. 2022 Aug;47(8):689-698. doi: 10.1016/j.tibs.2022.03.011. Epub 2022 Apr 6. Trends Biochem Sci. 2022. PMID: 35397924 Free PMC article. Review.
-
H2S and reactive sulfur signaling at the host-bacterial pathogen interface.J Biol Chem. 2020 Sep 18;295(38):13150-13168. doi: 10.1074/jbc.REV120.011304. Epub 2020 Jul 22. J Biol Chem. 2020. PMID: 32699012 Free PMC article. Review.
-
Emerging Chemical Biology of Protein Persulfidation.Antioxid Redox Signal. 2023 Jul;39(1-3):19-39. doi: 10.1089/ars.2023.0352. Epub 2023 Jul 10. Antioxid Redox Signal. 2023. PMID: 37288744 Free PMC article. Review.
References
-
- Argyrou A, and Blanchard JS (2004). Flavoprotein disulfide reductases: advances in chemistry and function. Progress in nucleic acid research and molecular biology 78, 89–142. - PubMed
-
- Babidge W, Millard S, and Roediger W (1998). Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: implications for ulcerative colitis. Mol. Cell. Biochem 181, 117–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

