HAMLET, a protein complex from human milk has bactericidal activity and enhances the activity of antibiotics against pathogenic Streptococci
- PMID: 31591115
- PMCID: PMC6879259
- DOI: 10.1128/AAC.01193-19
HAMLET, a protein complex from human milk has bactericidal activity and enhances the activity of antibiotics against pathogenic Streptococci
Abstract
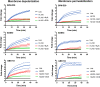
HAMLET is a protein-lipid complex derived from human milk that was first described for its tumoricidal activity. Later studies showed that HAMLET also has direct bactericidal activity against select species of bacteria, with highest activity against Streptococcus pneumoniae Additionally, HAMLET in combination with various antimicrobial agents can make a broader range of antibiotic-resistant bacterial species sensitive to antibiotics. Here, we show that HAMLET has direct antibacterial activity not only against pneumococci, but also against Streptococcus pyogenes (GAS) and Streptococcus agalactiae (GBS). Analogous to pneumococci, HAMLET-treatment of GAS and GBS resulted in depolarization of the bacterial membrane followed by membrane permeabilization and death that could be inhibited by calcium and sodium transport inhibitors. Treatment of clinical antibiotic-resistant isolates of S. pneumoniae, GAS, and GBS with sublethal concentrations of HAMLET in combination with antibiotics decreased the minimal inhibitory concentrations of the respective antibiotic into the sensitive range. This effect could also be blocked by ion transport inhibitors, suggesting that HAMLET's bactericidal and combination treatment effects used similar mechanisms. Finally, we show that HAMLET potentiated the effects of erythromycin against erythromycin-resistant bacteria more effectively than it potentiated killing by penicillin G of bacteria resistant to penicillin G. These results show for the first time that HAMLET effectively kills three different species of pathogenic Streptococci using similar mechanisms and also potentiate the activity of macrolides and lincosamides more effectively than combination treatment with beta-lactams. These findings suggest a potential therapeutic role for HAMLET in repurposing antibiotics currently causing treatment failures in patients.
Copyright © 2019 American Society for Microbiology.
Figures





References
-
- WHO. 2014. Antimicrobial resistance: global report of surveillance. WHO, Geneva, Switzerland.
-
- Tacconelli E, Carmeli Y, Harbarth S, Kahlmeter G, Kluytman J, Mendelson M, Pulcini C, Singh N, Theuretzbacker U. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO, Geneva, Switzerland.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

