Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs
- PMID: 31591196
- PMCID: PMC6815119
- DOI: 10.1073/pnas.1910613116
Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs
Abstract
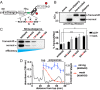
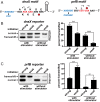
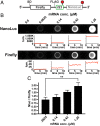
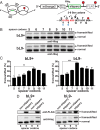
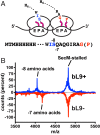
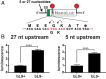
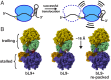
Translational frameshifting involves the repositioning of ribosomes on their messages into decoding frames that differ from those dictated during initiation. Some messenger RNAs (mRNAs) contain motifs that promote deliberate frameshifting to regulate production of the encoded proteins. The mechanisms of frameshifting have been investigated in many systems, and the resulting models generally involve single ribosomes responding to stimulator sequences in their engaged mRNAs. We discovered that the abundance of ribosomes on messages containing the IS3, dnaX, and prfB frameshift motifs significantly influences the levels of frameshifting. We show that this phenomenon results from ribosome collisions that occur during translational stalling, which can alter frameshifting in both the stalled and trailing ribosomes. Bacteria missing ribosomal protein bL9 are known to exhibit a reduction in reading frame maintenance and to have a strong dependence on elongation factor P (EFP). We discovered that ribosomes lacking bL9 become compacted closer together during collisions and that the E-sites of the stalled ribosomes appear to become blocked, which suggests subsequent transpeptidation in transiently stalled ribosomes may become compromised in the absence of bL9. In addition, we determined that bL9 can suppress frameshifting of its host ribosome, likely by regulating E-site dynamics. These findings provide mechanistic insight into the behavior of colliding ribosomes during translation and suggest naturally occurring frameshift elements may be regulated by the abundance of ribosomes relative to an mRNA pool.
Keywords: bL9; dnaX; frameshift; ribosome; translation.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
The energy landscape of -1 ribosomal frameshifting.Sci Adv. 2020 Jan 1;6(1):eaax6969. doi: 10.1126/sciadv.aax6969. eCollection 2020 Jan. Sci Adv. 2020. PMID: 31911945 Free PMC article.
-
Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2.Cell. 2004 Jul 9;118(1):45-55. doi: 10.1016/j.cell.2004.06.012. Cell. 2004. PMID: 15242643
-
Dynamic pathways of -1 translational frameshifting.Nature. 2014 Aug 21;512(7514):328-32. doi: 10.1038/nature13428. Epub 2014 Jun 11. Nature. 2014. PMID: 24919156 Free PMC article.
-
Ribosomal frameshifting, jumping and readthrough.Curr Opin Cell Biol. 1991 Dec;3(6):1051-5. doi: 10.1016/0955-0674(91)90128-l. Curr Opin Cell Biol. 1991. PMID: 1814364 Free PMC article. Review.
-
Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting.Trends Biochem Sci. 2015 May;40(5):265-74. doi: 10.1016/j.tibs.2015.03.006. Epub 2015 Apr 4. Trends Biochem Sci. 2015. PMID: 25850333 Free PMC article. Review.
Cited by
-
The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services.Nat Commun. 2023 Feb 25;14(1):1095. doi: 10.1038/s41467-023-36742-3. Nat Commun. 2023. PMID: 36841832 Free PMC article.
-
Ribosome-associated quality-control mechanisms from bacteria to humans.Mol Cell. 2022 Apr 21;82(8):1451-1466. doi: 10.1016/j.molcel.2022.03.038. Mol Cell. 2022. PMID: 35452614 Free PMC article. Review.
-
A cellular handbook for collided ribosomes: surveillance pathways and collision types.Curr Genet. 2021 Feb;67(1):19-26. doi: 10.1007/s00294-020-01111-w. Epub 2020 Oct 12. Curr Genet. 2021. PMID: 33044589 Free PMC article. Review.
-
Ribosome profiling of porcine reproductive and respiratory syndrome virus reveals novel features of viral gene expression.Elife. 2022 Feb 28;11:e75668. doi: 10.7554/eLife.75668. Elife. 2022. PMID: 35226596 Free PMC article.
-
The human mitochondrial mRNA structurome reveals mechanisms of gene expression.Science. 2024 Jul 19;385(6706):eadm9238. doi: 10.1126/science.adm9238. Epub 2024 Jul 19. Science. 2024. PMID: 39024447 Free PMC article.
References
-
- Rozov A., Demeshkina N., Westhof E., Yusupov M., Yusupova G., New structural insights into translational miscoding. Trends Biochem. Sci. 41, 798–814 (2016). - PubMed
-
- Chang B., Halgamuge S., Tang S. L., Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373, 90–99 (2006). - PubMed
-
- Studer S. M., Joseph S., Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 22, 105–115 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

