Regulation of human trophoblast syncytialization by histone demethylase LSD1
- PMID: 31591264
- PMCID: PMC6873176
- DOI: 10.1074/jbc.RA119.010518
Regulation of human trophoblast syncytialization by histone demethylase LSD1
Abstract
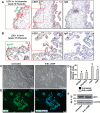
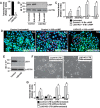
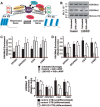
A successful pregnancy is critically dependent upon proper placental development and function. During human placentation, villous cytotrophoblast (CTB) progenitors differentiate to form syncytiotrophoblasts (SynTBs), which provide the exchange surface between the mother and fetus and secrete hormones to ensure proper progression of pregnancy. However, epigenetic mechanisms that regulate SynTB differentiation from CTB progenitors are incompletely understood. Here, we show that lysine-specific demethylase 1 (LSD1; also known as KDM1A), a histone demethylase, is essential to this process. LSD1 is expressed both in CTB progenitors and differentiated SynTBs in first-trimester placental villi; accordingly, expression in SynTBs is maintained throughout gestation. Impairment of LSD1 function in trophoblast progenitors inhibits induction of endogenous retrovirally encoded genes SYNCYTIN1/endogenous retrovirus group W member 1, envelope (ERVW1) and SYNCYTIN2/endogenous retrovirus group FRD member 1, envelope (ERVFRD1), encoding fusogenic proteins critical to human trophoblast syncytialization. Loss of LSD1 also impairs induction of chorionic gonadotropin α (CGA) and chorionic gonadotropin β (CGB) genes, which encode α and β subunits of human chorionic gonadotrophin (hCG), a hormone essential to modulate maternal physiology during pregnancy. Mechanistic analyses at the endogenous ERVW1, CGA, and CGB loci revealed a regulatory axis in which LSD1 induces demethylation of repressive histone H3 lysine 9 dimethylation (H3K9Me2) and interacts with transcription factor GATA2 to promote RNA polymerase II (RNA-POL-II) recruitment and activate gene transcription. Our study reveals a novel LSD1-GATA2 axis, which regulates human trophoblast syncytialization.
Keywords: LSD1; cytotrophoblast; histone demethylase; human; placenta; syncytiotrophoblast; transcription regulation; trophoblast.
© 2019 Milano-Foster et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

