Nucleoside Diphosphate Kinase Escalates A-to-C Mutations in MutT-Deficient Strains of Escherichia coli
- PMID: 31591275
- PMCID: PMC6932243
- DOI: 10.1128/JB.00567-19
Nucleoside Diphosphate Kinase Escalates A-to-C Mutations in MutT-Deficient Strains of Escherichia coli
Abstract
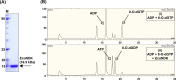
The chemical integrity of the nucleotide pool and its homeostasis are crucial for genome stability. Nucleoside diphosphate kinase (NDK) is a crucial enzyme that carries out reversible conversions from nucleoside diphosphate (NDP) to nucleoside triphosphate (NTP) and deoxynucleoside diphosphate (dNDP) to deoxynucleoside triphosphate (dNTP). Guanosine nucleotides (GDP, GTP, dGDP, and dGTP) are highly susceptible to oxidative damage to 8-oxo-GDP (8-O-GDP), 8-O-dGTP, 8-O-GTP, and 8-O-dGTP. MutT proteins in cells hydrolyze 8-O-GTP to 8-O-GMP or 8-O-dGTP to 8-O-dGMP to avoid its incorporation in nucleic acids. In Escherichia coli, 8-O-dGTP is also known to be hydrolyzed by RibA (GTP cyclohydrolase II). In this study, we show that E. coli NDK catalyzes the conversion of 8-O-dGDP to 8-O-dGTP or vice versa. However, the rate of NDK-mediated phosphorylation of 8-O-dGDP to 8-O-dGTP is about thrice as efficient as the rate of dephosphorylation of 8-O-dGTP to 8-O-dGDP, suggesting an additive role of NDK in net production of 8-O-dGTP in cells. Consistent with this observation, the depletion of NDK (Δndk) in E. coli ΔmutT or ΔmutT ΔribA strains results in a decrease of A-to-C mutations. These observations suggest that NDK contributes to the physiological load of MutT in E. coliIMPORTANCE Nucleoside diphosphate kinase (NDK), a ubiquitous enzyme, is known for its critical role in homeostasis of cellular nucleotide pools. However, NDK has now emerged as a molecule with pleiotropic effects in DNA repair, protein phosphorylation, gene expression, tumor metastasis, development, and pathogen virulence and persistence inside the host. In this study, we reveal an unexpected role of NDK in genome instability because of its activity in converting 8-O-dGDP to 8-O-dGTP. This observation has important consequences in escalating A-to-C mutations in Escherichia coli The severity of NDK in enhancing these mutations may be higher in the organisms challenged with high oxidative stress, which promotes 8-O-dGDP/8-O-dGTP production.
Keywords: 8-oxo-dGTP (8-O-dGTP); MutT; NDK; dNTP; mutation rate and mutation frequency.
Copyright © 2019 American Society for Microbiology.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

