Silencing Lysine-Specific Histone Demethylase 1 (LSD1) Causes Increased HP1-Positive Chromatin, Stimulation of DNA Repair Processes, and Dysregulation of Proliferation by Chk1 Phosphorylation in Human Endothelial Cells
- PMID: 31591366
- PMCID: PMC6829479
- DOI: 10.3390/cells8101212
Silencing Lysine-Specific Histone Demethylase 1 (LSD1) Causes Increased HP1-Positive Chromatin, Stimulation of DNA Repair Processes, and Dysregulation of Proliferation by Chk1 Phosphorylation in Human Endothelial Cells
Abstract
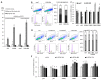
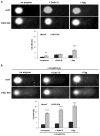
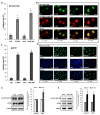
: The methylation of histone lysine residues modifies chromatin conformation and regulates the expression of genes implicated in cell metabolism. Lysine-specific demethylase 1 (LSD1) is a flavin-dependent monoamine oxidase that can demethylate mono- and dimethylated histone lysines 4 and 9 (H3K4 and H3K9). The removal of methyl groups from the lysine residues of histone and non-histone proteins was found to be an important regulatory factor of cell proliferation. However, its role has not been fully elucidated. In this study, we assessed LSD1-mediated cell cycle progression using a human endothelial cell model. The short hairpin RNA knockdown of LSD1 inhibits the G2/M phase of cell cycle progression by checkpoint kinase 1 (Chk1) phosphorylation (S137). We observed elevated DNA damage, which was consistent with the increased detection of double-strand breaks as well as purines and pyrimidines oxidation, which accompanied the activation of ATR/ATRIP signaling by H2AXS139 phosphorylation. The irreversible pharmacological inhibition of LSD1 by 2-phenylcyclopropylamine (2-PCPA) inactivated its enzymatic activity, causing significant changes in heterochromatin and euchromatin conformation assessed by chromatin assembly factor 1 subunit A (CAF1A) and heterochromatin protein 1 isoform α and γ (HP1α/γ) immunofluorescence analysis. We conclude that the knockdown of LSD1 in endothelial cells leads to increased HP1-positive chromatin, the stimulation of DNA repair processes, and the dysregulation of proliferation machinery.
Keywords: Chk1; DNA damage and repair; LSD1; cell cycle; chromatin remodeling; histone posttranslational modifications.
Conflict of interest statement
Funding: This research was funded by the Polish National Science Centre, project grant number: OPUS NCN 2012/05/B/NZ2/01663 and PRELUDIUM 2016/23/N/NZ3/02435.
Figures



Similar articles
-
Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4.Nature. 2010 Apr 1;464(7289):792-6. doi: 10.1038/nature08839. Epub 2010 Mar 14. Nature. 2010. PMID: 20228790
-
LSD1 promotes S-phase entry and tumorigenesis via chromatin co-occupation with E2F1 and selective H3K9 demethylation.Oncogene. 2018 Jan 25;37(4):534-543. doi: 10.1038/onc.2017.353. Epub 2017 Oct 9. Oncogene. 2018. PMID: 28991226
-
Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1β-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes.Arthritis Res Ther. 2014 May 16;16(3):R113. doi: 10.1186/ar4564. Arthritis Res Ther. 2014. PMID: 24886859 Free PMC article.
-
New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin.FEBS J. 2009 Aug;276(16):4304-12. doi: 10.1111/j.1742-4658.2009.07142.x. Epub 2009 Jul 14. FEBS J. 2009. PMID: 19624733 Review.
-
LSD1: more than demethylation of histone lysine residues.Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2. Epub 2020 Dec 14. Exp Mol Med. 2020. PMID: 33318631 Free PMC article. Review.
Cited by
-
Hydroxyurea and Caffeine Impact pRb-like Protein-Dependent Chromatin Architecture Profiles in Interphase Cells of Vicia faba.Int J Mol Sci. 2021 Apr 27;22(9):4572. doi: 10.3390/ijms22094572. Int J Mol Sci. 2021. PMID: 33925461 Free PMC article.
-
Genetic Modulation of the Erythrocyte Phenotype Associated with Retinopathy of Prematurity-A Multicenter Portuguese Cohort Study.Int J Mol Sci. 2023 Jul 23;24(14):11817. doi: 10.3390/ijms241411817. Int J Mol Sci. 2023. PMID: 37511576 Free PMC article.
-
LSD1 for the Targeted Regulation of Adipose Tissue.Curr Issues Mol Biol. 2022 Dec 27;45(1):151-163. doi: 10.3390/cimb45010012. Curr Issues Mol Biol. 2022. PMID: 36661498 Free PMC article. Review.
-
Kinetics of DNA Repair in Vicia faba Meristem Regeneration Following Replication Stress.Cells. 2021 Jan 7;10(1):88. doi: 10.3390/cells10010088. Cells. 2021. PMID: 33430297 Free PMC article.
-
Histone Methylation Related Therapeutic Challenge in Cardiovascular Diseases.Front Cardiovasc Med. 2021 Sep 9;8:710053. doi: 10.3389/fcvm.2021.710053. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34568453 Free PMC article. Review.
References
-
- Jin L., Hanigan C.L., Wu Y., Wang W., Park B.H., Woster P.M., Casero R.A. Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53- and DNMT1(DNA methyltransferase 1)-independent manner. Biochem. J. 2013;449:459–468. doi: 10.1042/BJ20121360. - DOI - PMC - PubMed
-
- Hille R., Miller S., Palfey B. Handbook of flavoproteins. De Gruyter; Berlin, Germany: 2013. pp. 119–138.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

