Integrating pharmacogenomics into the electronic health record by implementing genomic indicators
- PMID: 31591640
- PMCID: PMC6913212
- DOI: 10.1093/jamia/ocz177
Integrating pharmacogenomics into the electronic health record by implementing genomic indicators
Abstract
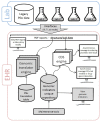
Pharmacogenomics (PGx) clinical decision support integrated into the electronic health record (EHR) has the potential to provide relevant knowledge to clinicians to enable individualized care. However, past experience implementing PGx clinical decision support into multiple EHR platforms has identified important clinical, procedural, and technical challenges. Commercial EHRs have been widely criticized for the lack of readiness to implement precision medicine. Herein, we share our experiences and lessons learned implementing new EHR functionality charting PGx phenotypes in a unique repository, genomic indicators, instead of using the problem or allergy list. The Gen-Ind has additional features including a brief description of the clinical impact, a hyperlink to the original laboratory report, and links to additional educational resources. The automatic generation of genomic indicators from interfaced PGx test results facilitates implementation and long-term maintenance of PGx data in the EHR and can be used as criteria for synchronous and asynchronous CDS.
Keywords: clinical decision support systems; delivery of health care; electronic health record; medical informatics; medication therapy management; pharmacogenetics; precision medicine.
© The Author(s) 2019. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Weinshilboum RM, Wang L.. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genom Hum Genet 2006; 7 (1): 223–45. - PubMed
-
- Clinical Pharmacogenetics Implementation Consortium (CPIC). 2019. https://cpicpgx.org/ Accessed May 6, 2019.
-
- Electronic Medical Records and Genomics (eMERGE) network. 2019. https://emerge.mc.vanderbilt.edu/ Accessed May 6, 2019.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

