Soft windowing application to improve analysis of high-throughput phenotyping data
- PMID: 31591642
- PMCID: PMC7115897
- DOI: 10.1093/bioinformatics/btz744
Soft windowing application to improve analysis of high-throughput phenotyping data
Abstract
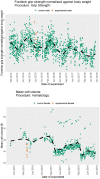
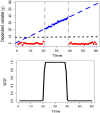
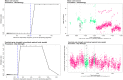
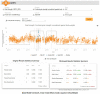
Motivation: High-throughput phenomic projects generate complex data from small treatment and large control groups that increase the power of the analyses but introduce variation over time. A method is needed to utlize a set of temporally local controls that maximizes analytic power while minimizing noise from unspecified environmental factors.
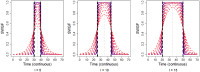
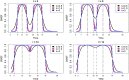
Results: Here we introduce 'soft windowing', a methodological approach that selects a window of time that includes the most appropriate controls for analysis. Using phenotype data from the International Mouse Phenotyping Consortium (IMPC), adaptive windows were applied such that control data collected proximally to mutants were assigned the maximal weight, while data collected earlier or later had less weight. We applied this method to IMPC data and compared the results with those obtained from a standard non-windowed approach. Validation was performed using a resampling approach in which we demonstrate a 10% reduction of false positives from 2.5 million analyses. We applied the method to our production analysis pipeline that establishes genotype-phenotype associations by comparing mutant versus control data. We report an increase of 30% in significant P-values, as well as linkage to 106 versus 99 disease models via phenotype overlap with the soft-windowed and non-windowed approaches, respectively, from a set of 2082 mutant mouse lines. Our method is generalizable and can benefit large-scale human phenomic projects such as the UK Biobank and the All of Us resources.
Availability and implementation: The method is freely available in the R package SmoothWin, available on CRAN http://CRAN.R-project.org/package=SmoothWin.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2019. Published by Oxford University Press.
Figures
References
-
- Begley C.G., Ellis L.M. (2012) Drug development: raise standards for preclinical cancer research. Nature, 483, 531–533. - PubMed

