Improved characterisation of MRSA transmission using within-host bacterial sequence diversity
- PMID: 31591959
- PMCID: PMC6954020
- DOI: 10.7554/eLife.46402
Improved characterisation of MRSA transmission using within-host bacterial sequence diversity
Abstract
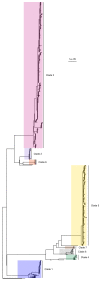
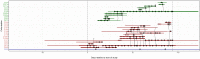
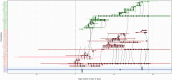
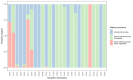
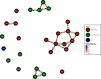
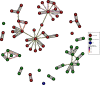
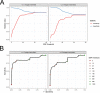
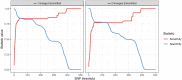
Methicillin-resistant Staphylococcus aureus (MRSA) transmission in the hospital setting has been a frequent subject of investigation using bacterial genomes, but previous approaches have not yet fully utilised the extra deductive power provided when multiple pathogen samples are acquired from each host. Here, we used a large dataset of MRSA sequences from multiply-sampled patients to reconstruct colonisation of individuals in a high-transmission setting in a hospital in Thailand. We reconstructed transmission trees for MRSA. We also investigated transmission between anatomical sites on the same individual, finding that this either occurs repeatedly or involves a wide transmission bottleneck. We examined the between-subject bottleneck, finding considerable variation in the amount of diversity transmitted. Finally, we compared our approach to the simpler method of identifying transmission pairs using single nucleotide polymorphism (SNP) counts. This suggested that the optimum threshold for identifying a pair is 39 SNPs, if sensitivities and specificities are equally weighted.
Keywords: MRSA; epidemiology; global health; hospital infections; human; molecular epidemiology.
© 2019, Hall et al.
Conflict of interest statement
MH, MH, PS, WM, VW, DL, KF, JP, EN, SP, CF No competing interests declared
Figures








References
-
- Azarian T, Cook RL, Johnson JA, Guzman N, McCarter YS, Gomez N, Rathore MH, Morris JG, Salemi M. Whole-genome sequencing for outbreak investigations of methicillin-resistant Staphylococcus aureus in the neonatal intensive care unit: time for routine practice? Infection Control & Hospital Epidemiology. 2015;36:777–785. doi: 10.1017/ice.2015.73. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

