DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism
- PMID: 31594538
- PMCID: PMC6781351
- DOI: 10.1186/s13072-019-0302-9
DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism
Abstract
Background: The stability of p53 is mainly controlled by ubiquitin-dependent degradation, which is triggered by the E3 ubiquitin ligase MDM2. The chromatin modifier lymphoid-specific helicase (LSH) is essential for DNA methylation and cancer progression as a transcriptional repressor. The potential interplay between chromatin modifiers and transcription factors remains largely unknown.
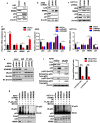
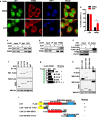
Results: Here, we present data suggesting that LSH regulates p53 in cis through two pathways: prevention proteasomal degradation through its deubiquitination, which is achieved by reducing the lysine 11-linked, lysine 48-linked polyubiquitin chains (K11 and K48) on p53; and revival of the transcriptional activity of p53 by forming a complex with PKM2 (pyruvate kinase 2). Furthermore, we confirmed that the LSH-PKM2 interaction occurred at the intersubunit interface region of the PKM2 C-terminal region and the coiled-coil domains (CC) and ATP-binding domains of LSH, and this interaction regulated p53-mediated transactivation in cis in lipid metabolism, especially lipid catabolism.
Conclusion: These findings suggest that LSH is a novel regulator of p53 through the proteasomal pathway, thereby providing an alternative mechanism of p53 involvement in lipid metabolism in cancer.
Keywords: DUB; LSH; Lipid metabolism; P53; PKM2.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Chromatin-Bound MDM2 Regulates Serine Metabolism and Redox Homeostasis Independently of p53.Mol Cell. 2016 Jun 16;62(6):890-902. doi: 10.1016/j.molcel.2016.04.033. Epub 2016 Jun 2. Mol Cell. 2016. PMID: 27264869
-
A role for LSH in facilitating DNA methylation by DNMT1 through enhancing UHRF1 chromatin association.Nucleic Acids Res. 2020 Dec 2;48(21):12116-12134. doi: 10.1093/nar/gkaa1003. Nucleic Acids Res. 2020. PMID: 33170271 Free PMC article.
-
TRIAD1 inhibits MDM2-mediated p53 ubiquitination and degradation.FEBS Lett. 2012 Sep 21;586(19):3057-63. doi: 10.1016/j.febslet.2012.07.022. Epub 2012 Jul 20. FEBS Lett. 2012. PMID: 22819825
-
Escape, or Vanish: Control the Fate of p53 through MDM2-Mediated Ubiquitination.Anticancer Agents Med Chem. 2015;16(2):174-89. doi: 10.2174/1871520615666150907093358. Anticancer Agents Med Chem. 2015. PMID: 26343143 Review.
-
The p53-Mdm2 module and the ubiquitin system.Semin Cancer Biol. 2003 Feb;13(1):49-58. doi: 10.1016/s1044-579x(02)00099-8. Semin Cancer Biol. 2003. PMID: 12507556 Review.
Cited by
-
The Organelle-Specific Regulations and Epigenetic Regulators in Ferroptosis.Front Pharmacol. 2022 Jun 17;13:905501. doi: 10.3389/fphar.2022.905501. eCollection 2022. Front Pharmacol. 2022. PMID: 35784729 Free PMC article. Review.
-
USP13 facilitates a ferroptosis-to-autophagy switch by activation of the NFE2L2/NRF2-SQSTM1/p62-KEAP1 axis dependent on the KRAS signaling pathway.Autophagy. 2025 Mar;21(3):565-582. doi: 10.1080/15548627.2024.2410619. Epub 2024 Oct 10. Autophagy. 2025. PMID: 39360581
-
High Glucose Induces Lipid Accumulation via 25-Hydroxycholesterol DNA-CpG Methylation.iScience. 2020 May 22;23(5):101102. doi: 10.1016/j.isci.2020.101102. Epub 2020 Apr 29. iScience. 2020. PMID: 32408171 Free PMC article.
-
The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression.Mol Cancer. 2020 Feb 27;19(1):39. doi: 10.1186/s12943-020-01157-x. Mol Cancer. 2020. PMID: 32103754 Free PMC article. Review.
-
Signaling pathways in cancer metabolism: mechanisms and therapeutic targets.Signal Transduct Target Ther. 2023 May 10;8(1):196. doi: 10.1038/s41392-023-01442-3. Signal Transduct Target Ther. 2023. PMID: 37164974 Free PMC article. Review.
References
-
- Prokesch A, Graef FA, Madl T, Kahlhofer J, Heidenreich S, Schumann A, Moyschewitz E, Pristoynik P, Blaschitz A, Knauer M, et al. Liver p53 is stabilized upon starvation and required for amino acid catabolism and gluconeogenesis. FASEB J. 2016;31(2):732–742. doi: 10.1096/fj.201600845R. - DOI - PMC - PubMed
-
- Liu Y, He Y, Jin A, Tikunov AP, Zhou L, Tollini LA, Leslie P, Kim TH, Li LO, Coleman RA, et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci USA. 2014;111(23):E2414–E2422. doi: 10.1073/pnas.1315605111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

