Plasmid Dissemination and Selection of a Multidrug-Resistant Klebsiella pneumoniae Strain during Transplant-Associated Antibiotic Therapy
- PMID: 31594809
- PMCID: PMC6786864
- DOI: 10.1128/mBio.00652-19
Plasmid Dissemination and Selection of a Multidrug-Resistant Klebsiella pneumoniae Strain during Transplant-Associated Antibiotic Therapy
Abstract
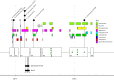
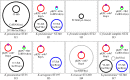
Antibiotics, which are used both to prevent and to treat infections, are a mainstay therapy for lifesaving procedures such as transplantation. For this reason, and many others, increased antibiotic resistance among human-associated pathogens, such as the carbapenem-resistant Enterobacteriaceae species, is of grave concern. In this study, we report on a hematopoietic stem cell transplant recipient in whom cultures detected the emergence of carbapenem resistance and spread across five strains of bacteria that persisted for over a year. Carbapenem resistance in Citrobacter freundii, Enterobacter cloacae, Klebsiella aerogenes, and Klebsiella pneumoniae was linked to a pair of plasmids, each carrying the Klebsiella pneumoniae carbapenemase gene (blaKPC). Surveillance cultures identified a carbapenem-susceptible strain of Citrobacter freundii that may have become resistant through horizontal gene transfer of these plasmids. Selection of a multidrug-resistant Klebsiella pneumoniae strain was also detected following combination antibiotic therapy. Here we report a plasmid carrying the blaKPC gene with broad host range that poses the additional threat of spreading to endogenous members of the human gut microbiome.IMPORTANCE Antibiotic-resistant bacteria are a serious threat to medically fragile patient populations. The spread of antibiotic resistance through plasmid-mediated mechanisms is of grave concern as it can lead to the conversion of endogenous patient-associated strains to difficult-to-treat pathogens.
Keywords: Enterobacteriaceae; HGT; Klebsiella; antibiotic resistance; carbapenems; genomes; plasmids.
Copyright © 2019 Conlan et al.
Figures

References
-
- Pecora N, Zhao X, Nudel K, Hoffmann M, Li N, Onderdonk AB, Yokoe D, Brown E, Allard M, Bry L. 2019. Diverse vectors and mechanisms spread New Delhi metallo-β-lactamases among carbapenem-resistant Enterobacteriaceae in the Greater Boston area. Antimicrob Agents Chemother 63:e02040-18. doi:10.1128/AAC.02040-18. - DOI - PMC - PubMed
-
- Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AKC, Carroll J, Scheld WM, Hazen KC, Sifri CD. 2011. Molecular dissection of an outbreak of carbapenem-resistant Enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. mBio 2:e00204-11. doi:10.1128/mBio.00204-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
