The secRNome of Listeria monocytogenes Harbors Small Noncoding RNAs That Are Potent Inducers of Beta Interferon
- PMID: 31594810
- PMCID: PMC6786865
- DOI: 10.1128/mBio.01223-19
The secRNome of Listeria monocytogenes Harbors Small Noncoding RNAs That Are Potent Inducers of Beta Interferon
Abstract
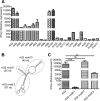
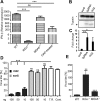
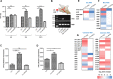
Cellular sensing of bacterial RNA is increasingly recognized as a determinant of host-pathogen interactions. The intracellular pathogen Listeria monocytogenes induces high levels of type I interferons (alpha/beta interferons [IFN-α/β]) to create a growth-permissive microenvironment during infection. We previously demonstrated that RNAs secreted by L. monocytogenes (comprising the secRNome) are potent inducers of IFN-β. We determined the composition and diversity of the members of the secRNome and found that they are uniquely enriched for noncoding small RNAs (sRNAs). Testing of individual sRNAs for their ability to induce IFN revealed several sRNAs with this property. We examined ril32, an intracellularly expressed sRNA that is highly conserved for the species L. monocytogenes and that was the most potent inducer of IFN-β expression of all the sRNAs tested in this study, in more detail. The rli32-induced IFN-β response is RIG-I (retinoic acid inducible gene I) dependent, and cells primed with rli32 inhibit influenza virus replication. We determined the rli32 motif required for IFN induction. rli32 overproduction promotes intracellular bacterial growth, and a mutant lacking rli32 is restricted for intracellular growth in macrophages. rli32-overproducing bacteria are resistant to H2O2 and exhibit both increased catalase activity and changes in the cell envelope. Comparative transcriptome sequencing (RNA-Seq) analysis indicated that ril32 regulates expression of the lhrC locus, previously shown to be involved in cell envelope stress. Inhibition of IFN-β signaling by ruxolitinib reduced rli32-dependent intracellular bacterial growth, indicating a link between induction of the interferon system and bacterial physiology. rli32 is, to the best of our knowledge, the first secreted individual bacterial sRNA known to trigger the induction of the type I IFN response.IMPORTANCE Interferons are potent and broadly acting cytokines that stimulate cellular responses to nucleic acids of unusual structures or locations. While protective when induced following viral infections, the induction of interferons is detrimental to the host during L. monocytogenes infection. Here, we identify specific sRNAs, secreted by the bacterium, with the capacity to induce type I IFN. Further analysis of the most potent sRNA, rli32, links the ability to induce RIG-I-dependent induction of the type I IFN response to the intracellular growth properties of the bacterium. Our findings emphasize the significance of released RNA for Listeria infection and shed light on a compartmental strategy used by an intracellular pathogen to modulate host responses to its advantage.
Keywords: Listeria monocytogenes; secreted RNA; type I IFN.
Copyright © 2019 Frantz et al.
Figures


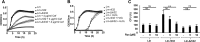
Similar articles
-
The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages.Nucleic Acids Res. 2011 May;39(10):4235-48. doi: 10.1093/nar/gkr033. Epub 2011 Jan 29. Nucleic Acids Res. 2011. PMID: 21278422 Free PMC article.
-
The multicopy sRNA LhrC controls expression of the oligopeptide-binding protein OppA in Listeria monocytogenes.RNA Biol. 2015;12(9):985-97. doi: 10.1080/15476286.2015.1071011. RNA Biol. 2015. PMID: 26176322 Free PMC article.
-
Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs.PLoS One. 2014 Feb 3;9(2):e83979. doi: 10.1371/journal.pone.0083979. eCollection 2014. PLoS One. 2014. PMID: 24498259 Free PMC article.
-
Identification and Role of Regulatory Non-Coding RNAs in Listeria monocytogenes.Int J Mol Sci. 2011;12(8):5070-9. doi: 10.3390/ijms12085070. Epub 2011 Aug 10. Int J Mol Sci. 2011. PMID: 21954346 Free PMC article. Review.
-
The non-coding RNA world of the bacterial pathogen Listeria monocytogenes.RNA Biol. 2012 Apr;9(4):372-8. doi: 10.4161/rna.19235. Epub 2012 Feb 16. RNA Biol. 2012. PMID: 22336762 Review.
Cited by
-
Interplay between Regulatory RNAs and Signal Transduction Systems during Bacterial Infection.Genes (Basel). 2020 Oct 16;11(10):1209. doi: 10.3390/genes11101209. Genes (Basel). 2020. PMID: 33081172 Free PMC article. Review.
-
Prevalence of small base-pairing RNAs derived from diverse genomic loci.Biochim Biophys Acta Gene Regul Mech. 2020 Jul;1863(7):194524. doi: 10.1016/j.bbagrm.2020.194524. Epub 2020 Mar 5. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32147527 Free PMC article. Review.
-
Nod-like Receptors: Critical Intracellular Sensors for Host Protection and Cell Death in Microbial and Parasitic Infections.Int J Mol Sci. 2021 Oct 22;22(21):11398. doi: 10.3390/ijms222111398. Int J Mol Sci. 2021. PMID: 34768828 Free PMC article. Review.
-
Extracellular Vesicles and Host-Pathogen Interactions: A Review of Inter-Kingdom Signaling by Small Noncoding RNA.Genes (Basel). 2021 Jun 30;12(7):1010. doi: 10.3390/genes12071010. Genes (Basel). 2021. PMID: 34208860 Free PMC article. Review.
-
Bacterial Membrane Vesicles: Physiological Roles, Infection Immunology, and Applications.Adv Sci (Weinh). 2023 Sep;10(25):e2301357. doi: 10.1002/advs.202301357. Epub 2023 Jun 25. Adv Sci (Weinh). 2023. PMID: 37357142 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

