Lotus japonicus Symbiosis Genes Impact Microbial Interactions between Symbionts and Multikingdom Commensal Communities
- PMID: 31594815
- PMCID: PMC6786870
- DOI: 10.1128/mBio.01833-19
Lotus japonicus Symbiosis Genes Impact Microbial Interactions between Symbionts and Multikingdom Commensal Communities
Abstract
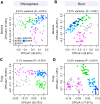
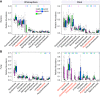
The wild legume Lotus japonicus engages in mutualistic symbiotic relationships with arbuscular mycorrhiza (AM) fungi and nitrogen-fixing rhizobia. Using plants grown in natural soil and community profiling of bacterial 16S rRNA genes and fungal internal transcribed spacers (ITSs), we examined the role of the Lotus symbiosis genes RAM1, NFR5, SYMRK, and CCaMK in structuring bacterial and fungal root-associated communities. We found host genotype-dependent community shifts in the root and rhizosphere compartments that were mainly confined to bacteria in nfr5 or fungi in ram1 mutants, while symrk and ccamk plants displayed major changes across both microbial kingdoms. We observed in all AM mutant roots an almost complete depletion of a large number of Glomeromycota taxa that was accompanied by a concomitant enrichment of Helotiales and Nectriaceae fungi, suggesting compensatory niche replacement within the fungal community. A subset of Glomeromycota whose colonization is strictly dependent on the common symbiosis pathway was retained in ram1 mutants, indicating that RAM1 is dispensable for intraradical colonization by some Glomeromycota fungi. However, intraradical colonization by bacteria belonging to the Burkholderiaceae and Anaeroplasmataceae is dependent on AM root infection, revealing a microbial interkingdom interaction. Despite the overall robustness of the bacterial root microbiota against major changes in the composition of root-associated fungal assemblages, bacterial and fungal cooccurrence network analysis demonstrates that simultaneous disruption of AM and rhizobium symbiosis increases the connectivity among taxa of the bacterial root microbiota. Our findings imply a broad role for Lotus symbiosis genes in structuring the root microbiota and identify unexpected microbial interkingdom interactions between root symbionts and commensal communities.IMPORTANCE Studies on symbiosis genes in plants typically focus on binary interactions between roots and soilborne nitrogen-fixing rhizobia or mycorrhizal fungi in laboratory environments. We utilized wild type and symbiosis mutants of a model legume, grown in natural soil, in which bacterial, fungal, or both symbioses are impaired to examine potential interactions between the symbionts and commensal microorganisms of the root microbiota when grown in natural soil. This revealed microbial interkingdom interactions between the root symbionts and fungal as well as bacterial commensal communities. Nevertheless, the bacterial root microbiota remains largely robust when fungal symbiosis is impaired. Our work implies a broad role for host symbiosis genes in structuring the root microbiota of legumes.
Keywords: microbiome; plant-microbe interactions; symbiosis.
Copyright © 2019 Thiergart et al.
Figures



Similar articles
-
Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases.Elife. 2014 Nov 25;3:e03891. doi: 10.7554/eLife.03891. Elife. 2014. PMID: 25422918 Free PMC article.
-
Dysfunction in the arbuscular mycorrhizal symbiosis has consistent but small effects on the establishment of the fungal microbiota in Lotus japonicus.New Phytol. 2019 Oct;224(1):409-420. doi: 10.1111/nph.15958. Epub 2019 Jul 2. New Phytol. 2019. PMID: 31125425 Free PMC article.
-
Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants.Mol Ecol. 2014 Jul;23(13):3356-70. doi: 10.1111/mec.12821. Mol Ecol. 2014. PMID: 24894495
-
Transcription factors network in root endosymbiosis establishment and development.World J Microbiol Biotechnol. 2018 Feb 15;34(3):37. doi: 10.1007/s11274-018-2418-7. World J Microbiol Biotechnol. 2018. PMID: 29450655 Review.
-
Arbuscular mycorrhizal fungi: a specialised niche for rhizospheric and endocellular bacteria.Antonie Van Leeuwenhoek. 2002 Aug;81(1-4):365-71. doi: 10.1023/a:1020544919072. Antonie Van Leeuwenhoek. 2002. PMID: 12448735 Review.
Cited by
-
Root microbiota assembly and adaptive differentiation among European Arabidopsis populations.Nat Ecol Evol. 2020 Jan;4(1):122-131. doi: 10.1038/s41559-019-1063-3. Epub 2019 Dec 23. Nat Ecol Evol. 2020. PMID: 31900452
-
Receptor-Like Kinases Sustain Symbiotic Scrutiny.Plant Physiol. 2020 Apr;182(4):1597-1612. doi: 10.1104/pp.19.01341. Epub 2020 Feb 13. Plant Physiol. 2020. PMID: 32054781 Free PMC article. Review.
-
Genome sequencing and functional analysis of a multipurpose medicinal herb Tinospora cordifolia (Giloy).Sci Rep. 2024 Feb 2;14(1):2799. doi: 10.1038/s41598-024-53176-z. Sci Rep. 2024. PMID: 38307917 Free PMC article.
-
Deciphering the rhizosphere microbiota composition of nature farming soybean (Glycine max L.) with different nodulation phenotypes.BMC Plant Biol. 2025 Apr 24;25(1):520. doi: 10.1186/s12870-025-06566-y. BMC Plant Biol. 2025. PMID: 40275151 Free PMC article.
-
A Eucalyptus Pht1 Family Gene EgPT8 Is Essential for Arbuscule Elongation of Rhizophagus irregularis.Microbiol Spectr. 2022 Dec 21;10(6):e0147022. doi: 10.1128/spectrum.01470-22. Epub 2022 Oct 13. Microbiol Spectr. 2022. PMID: 36227088 Free PMC article.
References
-
- Taylor TN, Remy W, Hass H, Kerp H. 1995. Fossil arbuscular mycorrhizae from the early Devonian. Mycologia 87:560–573. doi:10.1080/00275514.1995.12026569. - DOI
-
- Smith SE, Read DJ. 1997. Mycorrhizal symbiosis, 2nd ed, p 453–469. Academic Press, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

