Neuronal ER-plasma membrane junctions organized by Kv2-VAP pairing recruit Nir proteins and affect phosphoinositide homeostasis
- PMID: 31594866
- PMCID: PMC6879337
- DOI: 10.1074/jbc.RA119.007635
Neuronal ER-plasma membrane junctions organized by Kv2-VAP pairing recruit Nir proteins and affect phosphoinositide homeostasis
Abstract
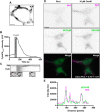
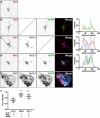
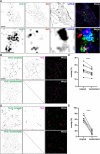
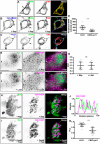
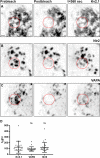
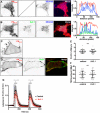
The association of plasma membrane (PM)-localized voltage-gated potassium (Kv2) channels with endoplasmic reticulum (ER)-localized vesicle-associated membrane protein-associated proteins VAPA and VAPB defines ER-PM junctions in mammalian brain neurons. Here, we used proteomics to identify proteins associated with Kv2/VAP-containing ER-PM junctions. We found that the VAP-interacting membrane-associated phosphatidylinositol (PtdIns) transfer proteins PYK2 N-terminal domain-interacting receptor 2 (Nir2) and Nir3 specifically associate with Kv2.1 complexes. When coexpressed with Kv2.1 and VAPA in HEK293T cells, Nir2 colocalized with cell-surface-conducting and -nonconducting Kv2.1 isoforms. This was enhanced by muscarinic-mediated PtdIns(4,5)P2 hydrolysis, leading to dynamic recruitment of Nir2 to Kv2.1 clusters. In cultured rat hippocampal neurons, exogenously expressed Nir2 did not strongly colocalize with Kv2.1, unless exogenous VAPA was also expressed, supporting the notion that VAPA mediates the spatial association of Kv2.1 and Nir2. Immunolabeling signals of endogenous Kv2.1, Nir2, and VAP puncta were spatially correlated in cultured neurons. Fluorescence-recovery-after-photobleaching experiments revealed that Kv2.1, VAPA, and Nir2 have comparable turnover rates at ER-PM junctions, suggesting that they form complexes at these sites. Exogenous Kv2.1 expression in HEK293T cells resulted in significant differences in the kinetics of PtdIns(4,5)P2 recovery following repetitive muscarinic stimulation, with no apparent impact on resting PtdIns(4,5)P2 or PtdIns(4)P levels. Finally, the brains of Kv2.1-knockout mice had altered composition of PtdIns lipids, suggesting a crucial role for native Kv2.1-containing ER-PM junctions in regulating PtdIns lipid metabolism in brain neurons. These results suggest that ER-PM junctions formed by Kv2 channel-VAP pairing regulate PtdIns lipid homeostasis via VAP-associated PtdIns transfer proteins.
Keywords: brain; endoplasmic reticulum (ER); lipids; membrane contact site (MCS); membrane protein; membrane-associated phosphatidylinositol transfer proteins; neuron; organelle; phosphatidylinositol signaling; plasma membrane; plasma membrane junction; potassium channel.
© 2019 Kirmiz et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

