TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging
- PMID: 31594908
- PMCID: PMC6781902
- DOI: 10.1534/genetics.119.302504
TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging
Abstract
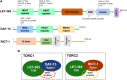
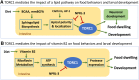
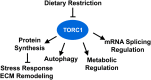
The Target of Rapamycin (TOR or mTOR) is a serine/threonine kinase that regulates growth, development, and behaviors by modulating protein synthesis, autophagy, and multiple other cellular processes in response to changes in nutrients and other cues. Over recent years, TOR has been studied intensively in mammalian cell culture and genetic systems because of its importance in growth, metabolism, cancer, and aging. Through its advantages for unbiased, and high-throughput, genetic and in vivo studies, Caenorhabditis elegans has made major contributions to our understanding of TOR biology. Genetic analyses in the worm have revealed unexpected aspects of TOR functions and regulation, and have the potential to further expand our understanding of how growth and metabolic regulation influence development. In the aging field, C. elegans has played a leading role in revealing the promise of TOR inhibition as a strategy for extending life span, and identifying mechanisms that function upstream and downstream of TOR to influence aging. Here, we review the state of the TOR field in C. elegans, and focus on what we have learned about its functions in development, metabolism, and aging. We discuss knowledge gaps, including the potential pitfalls in translating findings back and forth across organisms, but also describe how TOR is important for C. elegans biology, and how C. elegans work has developed paradigms of great importance for the broader TOR field.
Keywords: Caenorhabditis elegans development; DAF-15; NPRL-2; NPRL-3; Nprl2; Nprl3; RAGA-1; RSKS-1; RagA; RagC; Raptor; Rheb; Rheb-1; Rictor; S6 kinase; TOR; TORC1; TORC2; WormBook; aging; growth regulation; metabolism; nutrient signaling; sphingolipid.
Copyright © 2019 by the Genetics Society of America.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

