Spatiotemporal dynamics of multidrug resistant bacteria on intensive care unit surfaces
- PMID: 31594927
- PMCID: PMC6783542
- DOI: 10.1038/s41467-019-12563-1
Spatiotemporal dynamics of multidrug resistant bacteria on intensive care unit surfaces
Abstract
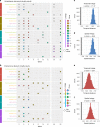
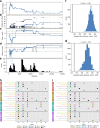
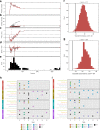
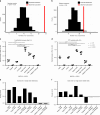
Bacterial pathogens that infect patients also contaminate hospital surfaces. These contaminants impact hospital infection control and epidemiology, prompting quantitative examination of their transmission dynamics. Here we investigate spatiotemporal and phylogenetic relationships of multidrug resistant (MDR) bacteria on intensive care unit surfaces from two hospitals in the United States (US) and Pakistan collected over one year. MDR bacteria isolated from 3.3% and 86.7% of US and Pakistani surfaces, respectively, include common nosocomial pathogens, rare opportunistic pathogens, and novel taxa. Common nosocomial isolates are dominated by single lineages of different clones, are phenotypically MDR, and have high resistance gene burdens. Many resistance genes (e.g., blaNDM, blaOXA carbapenamases), are shared by multiple species and flanked by mobilization elements. We identify Acinetobacter baumannii and Enterococcus faecium co-association on multiple surfaces, and demonstrate these species establish synergistic biofilms in vitro. Our results highlight substantial MDR pathogen burdens in hospital built-environments, provide evidence for spatiotemporal-dependent transmission, and demonstrate potential mechanisms for multi-species surface persistence.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Phenotypic and molecular characterization of Acinetobacter baumannii isolates causing lower respiratory infections among ICU patients.Microb Pathog. 2019 Mar;128:75-81. doi: 10.1016/j.micpath.2018.12.023. Epub 2018 Dec 15. Microb Pathog. 2019. PMID: 30562602
-
Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: a nationwide study.Ann Clin Microbiol Antimicrob. 2021 Mar 20;20(1):19. doi: 10.1186/s12941-021-00424-z. Ann Clin Microbiol Antimicrob. 2021. PMID: 33743736 Free PMC article.
-
Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients.BMC Infect Dis. 2019 Jul 17;19(1):629. doi: 10.1186/s12879-019-4272-0. BMC Infect Dis. 2019. PMID: 31315572 Free PMC article.
-
Emergence of antimicrobial resistance among Acinetobacter species: a global threat.Curr Opin Crit Care. 2016 Oct;22(5):491-9. doi: 10.1097/MCC.0000000000000337. Curr Opin Crit Care. 2016. PMID: 27552304 Review.
-
Acinetobacter baumannii: an emerging multidrug-resistant threat.Expert Rev Anti Infect Ther. 2008 Jun;6(3):309-25. doi: 10.1586/14787210.6.3.309. Expert Rev Anti Infect Ther. 2008. PMID: 18588496 Review.
Cited by
-
A systematic review and meta-analysis of carbapenem resistance and its possible treatment options with focus on clinical Enterobacteriaceae: Thirty years of development in Pakistan.Heliyon. 2024 Mar 17;10(7):e28052. doi: 10.1016/j.heliyon.2024.e28052. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38596009 Free PMC article.
-
Environmental Spread of Antibiotic Resistance.Antibiotics (Basel). 2021 May 27;10(6):640. doi: 10.3390/antibiotics10060640. Antibiotics (Basel). 2021. PMID: 34071771 Free PMC article. Review.
-
Biofilm Resilience: Molecular Mechanisms Driving Antibiotic Resistance in Clinical Contexts.Biology (Basel). 2025 Feb 6;14(2):165. doi: 10.3390/biology14020165. Biology (Basel). 2025. PMID: 40001933 Free PMC article. Review.
-
Microbial Air Quality in Healthcare Facilities.Int J Environ Res Public Health. 2021 Jun 9;18(12):6226. doi: 10.3390/ijerph18126226. Int J Environ Res Public Health. 2021. PMID: 34207509 Free PMC article. Review.
-
Commensal-pathogen dynamics structure disease outcomes during Clostridioides difficile colonization.Cell Host Microbe. 2025 Jan 8;33(1):30-41.e6. doi: 10.1016/j.chom.2024.12.002. Epub 2024 Dec 27. Cell Host Microbe. 2025. PMID: 39731916
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

