UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis
- PMID: 31595041
- PMCID: PMC6951344
- DOI: 10.1038/s41422-019-0236-6
UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis
Abstract
Protein biogenesis at the endoplasmic reticulum (ER) in eukaryotic cells is monitored by a protein quality control system named ER-associated protein degradation (ERAD). While there has been substantial progress in understanding how ERAD eliminates defective polypeptides generated from erroneous folding, how cells remove nascent chains stalled in the translocon during co-translational protein insertion into the ER is unclear. Here we show that ribosome stalling during protein translocation induces the attachment of UFM1, a ubiquitin-like modifier, to two conserved lysine residues near the COOH-terminus of the 60S ribosomal subunit RPL26 (uL24) at the ER. Strikingly, RPL26 UFMylation enables the degradation of stalled nascent chains, but unlike ERAD or previously established cytosolic ribosome-associated quality control (RQC), which uses proteasome to degrade their client proteins, ribosome UFMylation promotes the targeting of a translocation-arrested ER protein to lysosomes for degradation. RPL26 UFMylation is upregulated during erythroid differentiation to cope with increased secretory flow, and compromising UFMylation impairs protein secretion, and ultimately hemoglobin production. We propose that in metazoan, co-translational protein translocation into the ER is safeguarded by a UFMylation-dependent protein quality control mechanism, which when impaired causes anemia in mice and abnormal neuronal development in humans.
Conflict of interest statement
The authors declare no competing interests.
Figures

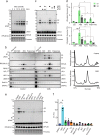

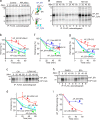
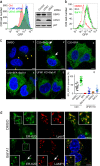

Comment in
-
Cleaning up stalled ribosome-translocon complexes with ufmylation.Cell Res. 2020 Jan;30(1):1-2. doi: 10.1038/s41422-019-0249-1. Cell Res. 2020. PMID: 31802008 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

