A robust 6-mRNA signature for prognosis prediction of pancreatic ductal adenocarcinoma
- PMID: 31595147
- PMCID: PMC6775308
- DOI: 10.7150/ijbs.32899
A robust 6-mRNA signature for prognosis prediction of pancreatic ductal adenocarcinoma
Abstract
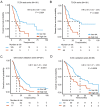
Pancreatic ductal adenocarcinoma (PDAC) is one of the most fatal malignancies worldwide. PDAC prognostic and diagnostic biomarkers are still being explored. The aim of this study is to establish a robust molecular signature that can improve the ability to predict PDAC prognosis. 155 overlapping differentially expressed genes between tumor and non-tumor tissues from three Gene Expression Omnibus (GEO) datasets were explored. A least absolute shrinkage and selection operator method (LASSO) Cox regression model was employed for selecting prognostic genes. We developed a 6-mRNA signature that can distinguish high PDAC risk patients from low risk patients with significant differences in overall survival (OS). We further validated this signature prognostic value in three independent cohorts (GEO batch, P < 0.0001; ICGC, P = 0.0036; Fudan, P = 0.029). Furthermore, we found that our signature remained significant in patients with different histologic grade, TNM stage, locations of tumor entity, age and gender. Multivariate cox regression analysis showed that 6-mRNA signature can be an independent prognostic marker in each of the cohorts. Receiver operating characteristic curve (ROC) analysis also showed that our signature possessed a better predictive role of PDAC prognosis. Moreover, the gene set enrichment analysis (GSEA) analysis showed that several tumorigenesis and metastasis related pathways were indeed associated with higher scores of risk. In conclusion, identifying the 6-mRNA signature could provide a valuable classification method to evaluate clinical prognosis and facilitate personalized treatment for PDAC patients. New therapeutic targets may be developed upon the functional analysis of the classifier genes and their related pathways.
Keywords: Pancreatic ductal adenocarcinoma; molecular signature; survival.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74:2913–21. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7–30. - PubMed
-
- Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet (London, England) 2016;388:73–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

