Enhanced GII.4 human norovirus infection in gnotobiotic pigs transplanted with a human gut microbiota
- PMID: 31596195
- PMCID: PMC7137776
- DOI: 10.1099/jgv.0.001336
Enhanced GII.4 human norovirus infection in gnotobiotic pigs transplanted with a human gut microbiota
Abstract
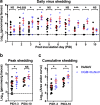
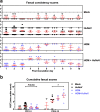
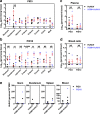
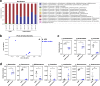
The role of commensal microbiota in enteric viral infections has been explored extensively, but the interaction between human gut microbiota (HGM) and human norovirus (HuNoV) is poorly understood. In this study, we established an HGM-Transplanted gnotobiotic (Gn) pig model of HuNoV infection and disease, using an infant stool as HGM transplant and a HuNoV GII.4/2006b strain for virus inoculation. Compared to germ-free Gn pigs, HuNoV inoculation in HGMT Gn pigs resulted in increased HuNoV shedding, characterized by significantly higher shedding titres on post inoculation day (PID) 3, 4, 6, 8 and 9, and significantly longer mean duration of virus shedding. In addition, virus titres were significantly higher in duodenum and distal ileum of HGMT Gn pigs on PID10, while comparable and transient HuNoV viremia was detected in both groups. 16S rRNA gene sequencing demonstrated that HuNoV infection dramatically altered intestinal microbiota in HGMT Gn pigs at the phylum (Proteobacteria, Firmicutes and Bacteroidetes) and genus (Enterococcus, Bifidobacterium, Clostridium, Ruminococcus, Anaerococcus, Bacteroides and Lactobacillus) levels. In summary, enhanced GII.4 HuNoV infection was observed in the presence of HGM, and host microbiota was susceptible to disruption upon HuNoV infection.
Keywords: Human norovirus; diarrhea; faecal microbiota transplantation; gnotobiotic pig; human gut microbiota.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

