Defective base excision repair in the response to DNA damaging agents in triple negative breast cancer
- PMID: 31596905
- PMCID: PMC6785058
- DOI: 10.1371/journal.pone.0223725
Defective base excision repair in the response to DNA damaging agents in triple negative breast cancer
Abstract
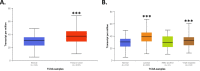
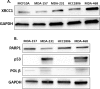
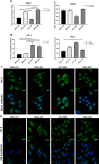
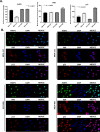
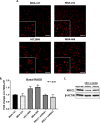
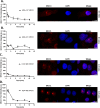
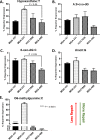
DNA repair defects have been increasingly focused on as therapeutic targets. In hormone-positive breast cancer, XRCC1-deficient tumors have been identified and proposed as targets for combination therapies that damage DNA and inhibit DNA repair pathways. XRCC1 is a scaffold protein that functions in base excision repair (BER) by mediating essential interactions between DNA glycosylases, AP endonuclease, poly(ADP-ribose) polymerase 1, DNA polymerase β (POL β), and DNA ligases. Loss of XRCC1 confers BER defects and hypersensitivity to DNA damaging agents. BER defects have not been evaluated in triple negative breast cancers (TNBC), for which new therapeutic targets and therapies are needed. To evaluate the potential of XRCC1 as an indicator of BER defects in TNBC, we examined XRCC1 expression in the TCGA database and its expression and localization in TNBC cell lines. The TCGA database revealed high XRCC1 expression in TNBC tumors and TNBC cell lines show variable, but mostly high expression of XRCC1. XRCC1 localized outside of the nucleus in some TNBC cell lines, altering their ability to repair base lesions and single-strand breaks. Subcellular localization of POL β also varied and did not correlate with XRCC1 localization. Basal levels of DNA damage correlated with observed changes in XRCC1 expression, localization, and measure repair capacity. The results confirmed that XRCC1 expression changes indicate DNA repair capacity changes but emphasize that basal DNA damage levels along with protein localization are better indicators of DNA repair defects. Given the observed over-expression of XRCC1 in TNBC preclinical models and tumors, XRCC1 expression levels should be assessed when evaluating treatment responses of TNBC preclinical model cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Wolf DM, Yau C, Sanil A, Glas A, Petricoin E, Wulfkuhle J, et al. DNA repair deficiency biomarkers and the 70-gene ultra-high risk signature as predictors of veliparib/carboplatin response in the I-SPY 2 breast cancer trial. NPJ Breast Cancer. 2017;3:31 10.1038/s41523-017-0025-7 - DOI - PMC - PubMed
-
- Abdel-Fatah TM, Russell R, Agarwal D, Moseley P, Abayomi MA, Perry C, et al. DNA polymerase beta deficiency is linked to aggressive breast cancer: a comprehensive analysis of gene copy number, mRNA and protein expression in multiple cohorts. Mol Oncol. 2014;8(3):520–32. 10.1016/j.molonc.2014.01.001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

