Illuminating biological pathways for drug targeting in head and neck squamous cell carcinoma
- PMID: 31596908
- PMCID: PMC6785123
- DOI: 10.1371/journal.pone.0223639
Illuminating biological pathways for drug targeting in head and neck squamous cell carcinoma
Abstract
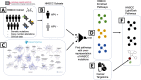
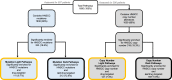
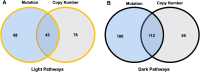
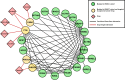
Head and neck squamous cell carcinoma (HNSCC) remains a morbid disease with poor prognosis and treatment that typically leaves patients with permanent damage to critical functions such as eating and talking. Currently only three targeted therapies are FDA approved for use in HNSCC, two of which are recently approved immunotherapies. In this work, we identify biological pathways involved with this disease that could potentially be targeted by current FDA approved cancer drugs and thereby expand the pool of potential therapies for use in HNSCC treatment. We analyzed 508 HNSCC patients with sequencing information from the Genomic Data Commons (GDC) database and assessed which biological pathways were significantly enriched for somatic mutations or copy number alterations. We then further classified pathways as either "light" or "dark" to the current reach of FDA-approved cancer drugs using the Cancer Targetome, a compendium of drug-target information. Light pathways are statistically enriched with somatic mutations (or copy number alterations) and contain one or more targets of current FDA-approved cancer drugs, while dark pathways are enriched with somatic mutations (or copy number alterations) but not currently targeted by FDA-approved cancer drugs. Our analyses indicated that approximately 35-38% of disease-specific pathways are in scope for repurposing of current cancer drugs. We further assess light and dark pathways for subgroups of patient tumor samples according to HPV status. The framework of light and dark pathways for HNSCC-enriched biological pathways allows us to better prioritize targeted therapies for further research in HNSCC based on the HNSCC genetic landscape and FDA-approved cancer drug information. We also highlight the importance in the identification of sub-pathways where targeting and cross targeting of other pathways may be most beneficial to predict positive or negative synergy with potential clinical significance. This framework is ideal for precision drug panel development, as well as identification of highly aberrant, untargeted candidates for future drug development.
Conflict of interest statement
Samuel Higgins is now employed by Roche Sequencing Solutions and Gabrielle Choonoo is now employed by Regeneron Pharmaceuticals. Their affiliation with these companies does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures

Similar articles
-
Exploiting high-throughput cell line drug screening studies to identify candidate therapeutic agents in head and neck cancer.BMC Pharmacol Toxicol. 2014 Nov 27;15:66. doi: 10.1186/2050-6511-15-66. BMC Pharmacol Toxicol. 2014. PMID: 25428177 Free PMC article.
-
Gene-expression signature regulated by the KEAP1-NRF2-CUL3 axis is associated with a poor prognosis in head and neck squamous cell cancer.BMC Cancer. 2018 Jan 6;18(1):46. doi: 10.1186/s12885-017-3907-z. BMC Cancer. 2018. PMID: 29306329 Free PMC article.
-
Emerging drugs for head and neck cancer.Expert Opin Emerg Drugs. 2015 Jun;20(2):313-29. doi: 10.1517/14728214.2015.1031653. Epub 2015 Mar 31. Expert Opin Emerg Drugs. 2015. PMID: 25826749 Free PMC article. Review.
-
Molecular pathways in head and neck cancer: EGFR, PI3K, and more.Am Soc Clin Oncol Educ Book. 2013:246-55. doi: 10.14694/EdBook_AM.2013.33.246. Am Soc Clin Oncol Educ Book. 2013. PMID: 23714515 Review.
-
Epidermal growth factor receptor (EGFR) and squamous cell carcinoma of the skin: molecular bases for EGFR-targeted therapy.Pathol Res Pract. 2011 Jun 15;207(6):337-42. doi: 10.1016/j.prp.2011.03.002. Epub 2011 Apr 29. Pathol Res Pract. 2011. PMID: 21531084 Review.
Cited by
-
Intrinsic Networks Regulating Tissue Repair: Comparative Studies of Oral and Skin Wound Healing.Cold Spring Harb Perspect Biol. 2022 Nov 1;14(11):a041244. doi: 10.1101/cshperspect.a041244. Cold Spring Harb Perspect Biol. 2022. PMID: 36041785 Free PMC article. Review.
-
Integrative analysis of drug response and clinical outcome in acute myeloid leukemia.Cancer Cell. 2022 Aug 8;40(8):850-864.e9. doi: 10.1016/j.ccell.2022.07.002. Epub 2022 Jul 21. Cancer Cell. 2022. PMID: 35868306 Free PMC article.
-
Contribution of Genomics to the Surgical Management and Study of Oral Cancer.Ann Surg Oncol. 2021 Oct;28(11):5842-5854. doi: 10.1245/s10434-021-09904-0. Epub 2021 Apr 12. Ann Surg Oncol. 2021. PMID: 33846893 Free PMC article. Review.
-
Assessing individual head and neck squamous cell carcinoma patient response to therapy through integration of functional and genomic data.Sci Rep. 2025 Jun 5;15(1):19742. doi: 10.1038/s41598-025-03111-7. Sci Rep. 2025. PMID: 40473660 Free PMC article.
References
-
- Union for International Cancer Control. Locally Advanced Squamous cell carcinoma of the head and neck [Internet]. World Health Organization; 2014. Available: https://www.who.int/selection_medicines/committees/expert/20/application...
-
- Cancer Facts & Figures 2018 [Internet]. American Cancer Society; 2018. Available: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-...
-
- Gillison ML, D’Souza G, Westra W, Sugar E, Xiao W, Begum S, et al. Distinct Risk Factor Profiles for Human Papillomavirus Type 16–Positive and Human Papillomavirus Type 16–Negative Head and Neck Cancers. JNCI: Journal of the National Cancer Institute. 2008;100: 407–420. 10.1093/jnci/djn025 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

