Chitosan Biosynthesis and Virulence in the Human Fungal Pathogen Cryptococcus gattii
- PMID: 31597720
- PMCID: PMC6796976
- DOI: 10.1128/mSphere.00644-19
Chitosan Biosynthesis and Virulence in the Human Fungal Pathogen Cryptococcus gattii
Abstract
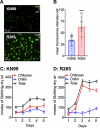
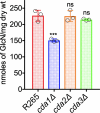
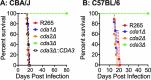
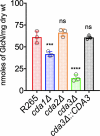
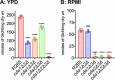
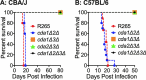
Cryptococcus gattii R265 is a hypervirulent fungal strain responsible for the recent outbreak of cryptococcosis in Vancouver Island of British Columbia in Canada. It differs significantly from Cryptococcus neoformans in its natural environment, its preferred site in the mammalian host, and its pathogenesis. Our previous studies of C. neoformans have shown that the presence of chitosan, the deacetylated form of chitin, in the cell wall attenuates inflammatory responses in the host, while its absence induces robust immune responses, which in turn facilitate clearance of the fungus and induces a protective response. The results of the present investigation reveal that the cell wall of C. gattii R265 contains a two- to threefold larger amount of chitosan than that of C. neoformans The genes responsible for the biosynthesis of chitosan are highly conserved in the R265 genome; the roles of the three chitin deacetylases (CDAs) have, however, been modified. To deduce their roles, single and double CDA deletion strains and a triple CDA deletion strain were constructed in a R265 background and were subjected to mammalian infection studies. Unlike C. neoformans where Cda1 has a discernible role in fungal pathogenesis, in strain R265, Cda3 is critical for virulence. Deletion of either CDA3 alone or in combination with another CDA (cda1Δ3Δ or cda2Δ3Δ) or both (cda1Δ2Δ3Δ) rendered the fungus avirulent and cleared from the infected host. Moreover, the cda1Δ2Δ3Δ strain of R265 induced a protective response to a subsequent infection with R265. These studies begin to illuminate the regulation of chitosan biosynthesis of C. gattii and its subsequent effect on fungal virulence.IMPORTANCE The fungal cell wall is an essential organelle whose components provide the first line of defense against host-induced antifungal activity. Chitosan is one of the carbohydrate polymers in the cell wall that significantly affects the outcome of host-pathogen interaction. Chitosan-deficient strains are avirulent, implicating chitosan as a critical virulence factor. C. gattii R265 is an important fungal pathogen of concern due to its ability to cause infections in individuals with no apparent immune dysfunction and an increasing geographical distribution. Characterization of the fungal cell wall and understanding the contribution of individual molecules of the cell wall matrix to fungal pathogenesis offer new therapeutic avenues for intervention. In this report, we show that the C. gattii R265 strain has evolved alternate regulation of chitosan biosynthesis under both laboratory growth conditions and during mammalian infection compared to that of C. neoformans.
Keywords: Cryptococcus gattii; R265; chitin; chitosan; chitosan regulation; protection; vaccine; virulence.
Copyright © 2019 Lam et al.
Figures


References
-
- Spickler AR. 2013. Cryptococcosis. The Center for Food Security and Public Health, Iowa State University, Ames, Iowa.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

