Isolation of Yasminevirus, the First Member of Klosneuvirinae Isolated in Coculture with Vermamoeba vermiformis, Demonstrates an Extended Arsenal of Translational Apparatus Components
- PMID: 31597770
- PMCID: PMC6912108
- DOI: 10.1128/JVI.01534-19
Isolation of Yasminevirus, the First Member of Klosneuvirinae Isolated in Coculture with Vermamoeba vermiformis, Demonstrates an Extended Arsenal of Translational Apparatus Components
Abstract
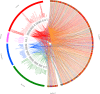
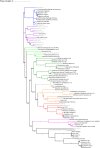
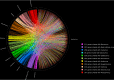
The family of giant viruses is still expanding, and evidence of a translational machinery is emerging in the virosphere. The Klosneuvirinae group of giant viruses was first reconstructed from in silico studies, and then a unique member was isolated, Bodo saltans virus. Here we describe the isolation of a new member in this group using coculture with the free-living amoeba Vermamoeba vermiformis This giant virus, called Yasminevirus, has a 2.1-Mb linear double-stranded DNA genome encoding 1,541 candidate proteins, with a GC content estimated at 40.2%. Yasminevirus possesses a nearly complete translational machinery, with a set of 70 tRNAs associated with 45 codons and recognizing 20 amino acids (aa), 20 aminoacyl-tRNA synthetases (aaRSs) recognizing 20 aa, as well as several translation factors and elongation factors. At the genome scale, evolutionary analyses placed this virus in the Klosneuvirinae group of giant viruses. Rhizome analysis demonstrated that the genome of Yasminevirus is mosaic, with ∼34% of genes having their closest homologues in other viruses, followed by ∼13.2% in Eukaryota, ∼7.2% in Bacteria, and less than 1% in Archaea Among giant virus sequences, Yasminevirus shared 87% of viral hits with Klosneuvirinae. This description of Yasminevirus sheds light on the Klosneuvirinae group in a captivating quest to understand the evolution and diversity of giant viruses.IMPORTANCE Yasminevirus is an icosahedral double-stranded DNA virus isolated from sewage water by amoeba coculture. Here its structure and replicative cycle in the amoeba Vermamoeba vermiformis are described and genomic and evolutionary studies are reported. This virus belongs to the Klosneuvirinae group of giant viruses, representing the second isolated and cultivated giant virus in this group, and is the first isolated using a coculture procedure. Extended translational machinery pointed to Yasminevirus among the quasiautonomous giant viruses with the most complete translational apparatus of the known virosphere.
Keywords: Klosneuvirinae; NCLDV; amoeba; giant virus; translation.
Copyright © 2019 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

