Yellow Fever Virus Reemergence and Spread in Southeast Brazil, 2016-2019
- PMID: 31597773
- PMCID: PMC6912119
- DOI: 10.1128/JVI.01623-19
Yellow Fever Virus Reemergence and Spread in Southeast Brazil, 2016-2019
Erratum in
-
Correction for Giovanetti et al., "Yellow Fever Virus Reemergence and Spread in Southeast Brazil, 2016-2019".J Virol. 2020 May 18;94(11):e02008-19. doi: 10.1128/JVI.02008-19. Print 2020 May 18. J Virol. 2020. PMID: 32423967 Free PMC article. No abstract available.
Abstract
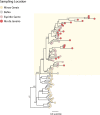
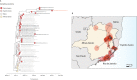
The recent reemergence of yellow fever virus (YFV) in Brazil has raised serious concerns due to the rapid dissemination of the virus in the southeastern region. To better understand YFV genetic diversity and dynamics during the recent outbreak in southeastern Brazil, we generated 18 complete and nearly complete genomes from the peak of the epidemic curve from nonhuman primates (NHPs) and human infected cases across the Espírito Santo and Rio de Janeiro states. Genomic sequencing of 18 YFV genomes revealed the estimated timing, source, and likely routes of yellow fever virus transmission and dispersion during one of the largest outbreaks ever registered in Brazil. We showed that during the recent epidemic, YFV was reintroduced from Minas Gerais to the Espírito Santo and Rio de Janeiro states multiple times between 2016 and 2019. The analysis of data from portable sequencing could identify the corridor of spread of YFV. These findings reinforce the idea that continued genomic surveillance strategies can provide information on virus genetic diversity and transmission dynamics that might assist in understanding arbovirus epidemics.IMPORTANCE Arbovirus infections in Brazil, including yellow fever, dengue, zika, and chikungunya, result in considerable morbidity and mortality and are pressing public health concerns. However, our understanding of these outbreaks is hampered by the limited availability of genomic data. In this study, we investigated the genetic diversity and spatial distribution of YFV during the current outbreak by analyzing genomic data from areas in southeastern Brazil not covered by other previous studies. To gain insights into the routes of YFV introduction and dispersion, we tracked the virus by sequencing YFV genomes sampled from nonhuman primates and infected patients from the southeastern region. Our study provides an understanding of how YFV initiates transmission in new Brazilian regions and illustrates that genomics in the field can augment traditional approaches to infectious disease surveillance and control.
Keywords: Southeast Brazil; genomic surveillance; outbreak; outbreak response; yellow fever.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Nunes MR, Palacios G, Cardoso JF, Martins LC, Sousa EC Jr, de Lima CP, Medeiros DB, Savji N, Desai A, Rodrigues SG, Carvalho VL, Lipkin WI, Vasconcelos PF. 2012. Genomic and phylogenetic characterization of Brazilian yellow fever virus strains. J Virol 86:13263–13271. doi: 10.1128/JVI.00565-12. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

