Evaluating the Intactness of Persistent Viral Genomes in Simian Immunodeficiency Virus-Infected Rhesus Macaques after Initiating Antiretroviral Therapy within One Year of Infection
- PMID: 31597776
- PMCID: PMC6912123
- DOI: 10.1128/JVI.01308-19
Evaluating the Intactness of Persistent Viral Genomes in Simian Immunodeficiency Virus-Infected Rhesus Macaques after Initiating Antiretroviral Therapy within One Year of Infection
Abstract
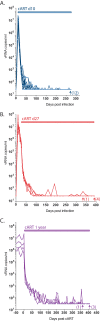
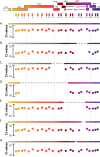
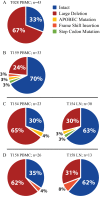
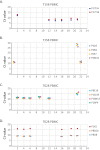
The major obstacle to more-definitive treatment for HIV infection is the early establishment of virus that persists despite long-term combination antiretroviral therapy (cART) and can cause recrudescent viremia if cART is interrupted. Previous studies of HIV DNA that persists despite cART indicated that only a small fraction of persistent viral sequences was intact. Experimental simian immunodeficiency virus (SIV) infections of nonhuman primates (NHPs) are essential models for testing interventions designed to reduce the viral reservoir. We studied the viral genomic integrity of virus that persists during cART under conditions typical of many NHP reservoir studies, specifically with cART started within 1 year postinfection and continued for at least 9 months. The fraction of persistent DNA in SIV-infected NHPs starting cART during acute or chronic infection was assessed with a multiamplicon, real-time PCR assay designed to analyze locations that are regularly spaced across the viral genome to maximize coverage (collectively referred to as "tile assay") combined with near-full-length (nFL) single-genome sequencing. The tile assay is used to rapidly screen for major deletions, with nFL sequence analysis used to identify additional potentially inactivating mutations. Peripheral blood mononuclear cells (PBMC) from animals started on cART within 1 month of infection, sampled at least 9 months after cART initiation, contained at least 80% intact genomes, whereas those from animals started on cART 1 year postinfection and treated for 1 year contained intact genomes only 47% of the time. The most common defect identified was large deletions, with the remaining defects caused by APOBEC-mediated mutations, frameshift mutations, and inactivating point mutations. Overall, this approach can be used to assess the intactness of persistent viral DNA in NHPs.IMPORTANCE Molecularly defining the viral reservoir that persists despite antiretroviral therapy and that can lead to rebound viremia if antiviral therapy is removed is critical for testing interventions aimed at reducing this reservoir. In HIV infection in humans with delayed treatment initiation and extended treatment duration, persistent viral DNA has been shown to be dominated by nonfunctional genomes. Using multiple real-time PCR assays across the genome combined with near-full-genome sequencing, we defined SIV genetic integrity after 9 to 18 months of combination antiretroviral therapy in rhesus macaques starting therapy within 1 year of infection. In the animals starting therapy within a month of infection, the vast majority of persistent DNA was intact and presumptively functional. Starting therapy within 1 year increased the nonintact fraction of persistent viral DNA. The approach described here allows rapid screening of viral intactness and is a valuable tool for assessing the efficacy of novel reservoir-reducing interventions.
Keywords: SIV; genome intactness; genomic integrity; persistent viral reservoir; single-genome sequencing.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. - DOI - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

