Spliceosomal disruption of the non-canonical BAF complex in cancer
- PMID: 31597964
- PMCID: PMC6858563
- DOI: 10.1038/s41586-019-1646-9
Spliceosomal disruption of the non-canonical BAF complex in cancer
Abstract
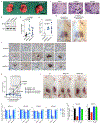
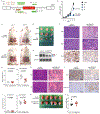
SF3B1 is the most commonly mutated RNA splicing factor in cancer1-4, but the mechanisms by which SF3B1 mutations promote malignancy are poorly understood. Here we integrated pan-cancer splicing analyses with a positive-enrichment CRISPR screen to prioritize splicing alterations that promote tumorigenesis. We report that diverse SF3B1 mutations converge on repression of BRD9, which is a core component of the recently described non-canonical BAF chromatin-remodelling complex that also contains GLTSCR1 and GLTSCR1L5-7. Mutant SF3B1 recognizes an aberrant, deep intronic branchpoint within BRD9 and thereby induces the inclusion of a poison exon that is derived from an endogenous retroviral element and subsequent degradation of BRD9 mRNA. Depletion of BRD9 causes the loss of non-canonical BAF at CTCF-associated loci and promotes melanomagenesis. BRD9 is a potent tumour suppressor in uveal melanoma, such that correcting mis-splicing of BRD9 in SF3B1-mutant cells using antisense oligonucleotides or CRISPR-directed mutagenesis suppresses tumour growth. Our results implicate the disruption of non-canonical BAF in the diverse cancer types that carry SF3B1 mutations and suggest a mechanism-based therapeutic approach for treating these malignancies.
Conflict of interest statement
The authors declare competing financial interests: details are available in the online version of the paper. Readers are welcome to comment on the online version of the paper.
Figures












References
-
- Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478, 64–69 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

