A general fluorescent light-up probe for staining and quantifying protein
- PMID: 31598246
- PMCID: PMC6731743
- DOI: 10.1098/rsos.190580
A general fluorescent light-up probe for staining and quantifying protein
Abstract
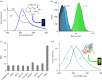
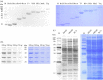
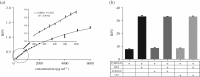
Proteins are the primary functional agents in all cellular processes, facilitating various functions such as enzymes and structure-forming or signal-transducing molecules. In this work, we report a fluorescent dye, PyMDI-Zn, which could specifically bind with proteins and provide a red-shifted fluorescent emission. The visual analysis of protein in sodium dodecyl sulfate-polyacrylamide gel electrophoresis could be realized in 5 min by using PyMDI-Zn as a light-up dye. Based on its cell penetration and low toxicity, PyMDI-Zn could also be applied to locate protein-rich regions and organelles in live cell imaging. Moreover, the direct protein quantitation can be realized based on PyMDI-Zn, providing a method of screening for food adulteration by nitrogen-rich compounds.
Keywords: PyMDI-Zn; SDS-PAGE; live cell imaging; protein quantitation.
© 2019 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures

Similar articles
-
Design and synthesis of a novel fluorescent protein probe for easy and rapid electrophoretic gel staining by using a commonly available UV-based fluorescent imaging system.Electrophoresis. 2013 Sep;34(17):2464-72. doi: 10.1002/elps.201300089. Electrophoresis. 2013. PMID: 23801451
-
Mixed anionic detergent/aliphatic alcohol-polyacrylamide gel electrophoresis alters the separation of proteins relative to conventional sodium dodecyl sulfate-polyacrylamide gel electrophoresis.Anal Biochem. 1988 Oct;174(1):337-48. doi: 10.1016/0003-2697(88)90555-6. Anal Biochem. 1988. PMID: 3218745
-
Use of the hydrophobic probe Nile red for the fluorescent staining of protein bands in sodium dodecyl sulfate-polyacrylamide gels.Anal Biochem. 1991 Dec;199(2):169-74. doi: 10.1016/0003-2697(91)90085-8. Anal Biochem. 1991. PMID: 1725949
-
Fluorescent labeling of proteins with nile red and 2-methoxy-2,4-diphenyl-3(2H)-furanone: physicochemical basis and application to the rapid staining of sodium dodecyl sulfate polyacrylamide gels and Western blots.Electrophoresis. 2001 Mar;22(5):874-80. doi: 10.1002/1522-2683()22:5<874::AID-ELPS874>3.0.CO;2-U. Electrophoresis. 2001. PMID: 11332755 Review.
-
The detection of enzyme activity following sodium dodecyl sulfate-polyacrylamide gel electrophoresis.Anal Biochem. 1998 Jun 15;260(1):1-17. doi: 10.1006/abio.1998.2680. Anal Biochem. 1998. PMID: 9648646 Review.
Cited by
-
Identification of Tunable, Environmentally Responsive Fluorogenic Dyes by High-Throughput Screening.ACS Chem Biol. 2024 Sep 20;19(9):2041-2049. doi: 10.1021/acschembio.4c00373. Epub 2024 Sep 9. ACS Chem Biol. 2024. PMID: 39250827 Free PMC article.
References
-
- Lodish H, Berk A, Matsudaira P, Kaiser CA, Kreiger M, Scott MP, Zipursky SL, Darnell J.. 2004. Molecular cell biology, 5th edn New York, NY: W. H. Freeman Company.
-
- Mathews CK, Van Holde KE, Ahern KG. 2000. Biochemistry, 3rd edn Reading, MA: Addison-Wesley Publishing Company.
Associated data
LinkOut - more resources
Full Text Sources

