Retrons and their applications in genome engineering
- PMID: 31598685
- PMCID: PMC6868368
- DOI: 10.1093/nar/gkz865
Retrons and their applications in genome engineering
Abstract
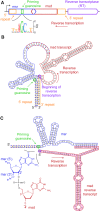
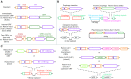
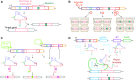
Precision genome editing technologies have transformed modern biology. These technologies have arisen from the redirection of natural biological machinery, such as bacteriophage lambda proteins for recombineering and CRISPR nucleases for eliciting site-specific double-strand breaks. Less well-known is a widely distributed class of bacterial retroelements, retrons, that employ specialized reverse transcriptases to produce noncoding intracellular DNAs. Retrons' natural function and mechanism of genetic transmission have remained enigmatic. However, recent studies have harnessed their ability to produce DNA in situ for genome editing and evolution. This review describes retron biology and function in both natural and synthetic contexts. We also highlight areas that require further study to advance retron-based precision genome editing platforms.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Greener A., Callahan M., Jerpseth B.. Trower MK. In Vitro Mutagenesis Protocols. 1996; Totowa: Humana Press; 375–385.
-
- Petri R., Schmidt-Dannert C.. Dealing with complexity: evolutionary engineering and genome shuffling. Curr. Opin. Biotechnol. 2004; 15:298–304. - PubMed
-
- Chen K., Arnold F.H.. Enzyme engineering for nonaqueous solvents: random mutagenesis to enhance activity of subtilisin E in polar organic media. Bio/Technology. 1991; 9:1073. - PubMed
-
- Crameri A., Raillard S.-A., Bermudez E., Stemmer W.P.C.. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998; 391:288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

