MirGeneDB 2.0: the metazoan microRNA complement
- PMID: 31598695
- PMCID: PMC6943042
- DOI: 10.1093/nar/gkz885
MirGeneDB 2.0: the metazoan microRNA complement
Erratum in
-
MirGeneDB 2.0: the metazoan microRNA complement.Nucleic Acids Res. 2020 Jan 8;48(D1):D1172. doi: 10.1093/nar/gkz1016. Nucleic Acids Res. 2020. PMID: 31642479 Free PMC article. No abstract available.
Abstract
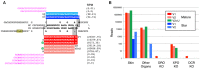
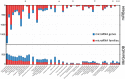
Small non-coding RNAs have gained substantial attention due to their roles in animal development and human disorders. Among them, microRNAs are special because individual gene sequences are conserved across the animal kingdom. In addition, unique and mechanistically well understood features can clearly distinguish bona fide miRNAs from the myriad other small RNAs generated by cells. However, making this distinction is not a common practice and, thus, not surprisingly, the heterogeneous quality of available miRNA complements has become a major concern in microRNA research. We addressed this by extensively expanding our curated microRNA gene database - MirGeneDB - to 45 organisms, encompassing a wide phylogenetic swath of animal evolution. By consistently annotating and naming 10,899 microRNA genes in these organisms, we show that previous microRNA annotations contained not only many false positives, but surprisingly lacked >2000 bona fide microRNAs. Indeed, curated microRNA complements of closely related organisms are very similar and can be used to reconstruct ancestral miRNA repertoires. MirGeneDB represents a robust platform for microRNA-based research, providing deeper and more significant insights into the biology and evolution of miRNAs as well as biomedical and biomarker research. MirGeneDB is publicly and freely available at http://mirgenedb.org/.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Matera A.G., Terns R.M., Terns M.P.. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007; 8:209–220. - PubMed
-
- Hamilton A.J., Baulcombe D.C.. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999; 286:950–952. - PubMed
-
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E.. Characterization of the piRNA complex from rat testes. Science. 2006; 313:363–367. - PubMed
-
- Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. - PubMed
-
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T.. Identification of novel genes coding for small expressed RNAs. Science. 2001; 294:853–858. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

