Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 3.0
- PMID: 31599596
- PMCID: PMC6986310
- DOI: 10.1021/acs.jproteome.9b00542
Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 3.0
Abstract
The Human Proteome Organization's (HUPO) Human Proteome Project (HPP) developed Mass Spectrometry (MS) Data Interpretation Guidelines that have been applied since 2016. These guidelines have helped ensure that the emerging draft of the complete human proteome is highly accurate and with low numbers of false-positive protein identifications. Here, we describe an update to these guidelines based on consensus-reaching discussions with the wider HPP community over the past year. The revised 3.0 guidelines address several major and minor identified gaps. We have added guidelines for emerging data independent acquisition (DIA) MS workflows and for use of the new Universal Spectrum Identifier (USI) system being developed by the HUPO Proteomics Standards Initiative (PSI). In addition, we discuss updates to the standard HPP pipeline for collecting MS evidence for all proteins in the HPP, including refinements to minimum evidence. We present a new plan for incorporating MassIVE-KB into the HPP pipeline for the next (HPP 2020) cycle in order to obtain more comprehensive coverage of public MS data sets. The main checklist has been reorganized under headings and subitems, and related guidelines have been grouped. In sum, Version 2.1 of the HPP MS Data Interpretation Guidelines has served well, and this timely update to version 3.0 will aid the HPP as it approaches its goal of collecting and curating MS evidence of translation and expression for all predicted ∼20 000 human proteins encoded by the human genome.
Keywords: B/D-HPP; C-HPP; HPP; Human Proteome Project; Universal Spectrum Identifier (USI); false-discovery rates; guidelines; mass spectrometry; standards; unicity checker.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
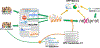
References
-
- Omenn GS; Lane L; Overall CM; Corrales FJ; Schwenk JM; Paik Y-K; Van Eyk JE; Liu S; Snyder M; Baker MS; et al. Progress on Identifying and Characterizing the Human Proteome: 2018 Metrics from the HUPO Human Proteome Project. J. Proteome Res 2018, 17 (12), 4031–4041. 10.1021/acs.jproteome.8b00441. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 GM127667/GM/NIGMS NIH HHS/United States
- U54 EB020406/EB/NIBIB NIH HHS/United States
- U19 AG023122/AG/NIA NIH HHS/United States
- P41 GM103484/GM/NIGMS NIH HHS/United States
- R01 GM087221/GM/NIGMS NIH HHS/United States
- R01 LM013115/LM/NLM NIH HHS/United States
- U24 HG007822/HG/NHGRI NIH HHS/United States
- P30 ES017885/ES/NIEHS NIH HHS/United States
- R01 HL111362/HL/NHLBI NIH HHS/United States
- U41 HG007822/HG/NHGRI NIH HHS/United States
- R01 HL133135/HL/NHLBI NIH HHS/United States
- U24 CA210967/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

