Doxycycline-Dependent Self-Inactivation of CRISPR-Cas9 to Temporally Regulate On- and Off-Target Editing
- PMID: 31601489
- PMCID: PMC6952177
- DOI: 10.1016/j.ymthe.2019.09.006
Doxycycline-Dependent Self-Inactivation of CRISPR-Cas9 to Temporally Regulate On- and Off-Target Editing
Abstract
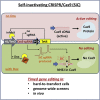
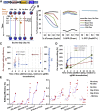
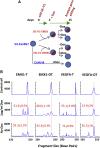
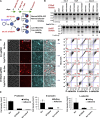
Exome and deep sequencing of cells treated with a panel of lentiviral guide RNA demonstrate that both on- and off-target editing proceed in a time-dependent manner. Thus, methods to temporally control Cas9 activity would be beneficial. To address this need, we describe a "self-inactivating CRISPR (SiC)" system consisting of a single guide RNA that deactivates the Streptococcus pyogenes Cas9 nuclease in a doxycycline-dependent manner. This enables defined, temporal control of Cas9 activity in any cell type and also in vivo. Results show that SiC may enable a reduction in off-target editing, with less effect on on-target editing rates. This tool facilitates diverse applications including (1) the timed regulation of genetic knockouts in hard-to-transfect cells using lentivirus, including human leukocytes for the identification of glycogenes regulating leukocyte-endothelial cell adhesion; (2) genome-wide lentiviral sgRNA (single guide RNA) library applications where Cas9 activity is ablated after allowing pre-determined editing times. Thus, stable knockout cell pools are created for functional screens; and (3) temporal control of Cas9-mediated editing of myeloid and lymphoid cells in vivo, both in mouse peripheral blood and bone marrow. Overall, SiC enables temporal control of gene editing and may be applied in diverse application including studies that aim to reduce off-target genome editing.
Keywords: CRISPR; Cas9; doxycycline; exome sequencing; genome editing; genome-wide screen; hematopoietic stem cells; lentivirus; next generation sequencing; off target editing.
Copyright © 2019 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

