ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation
- PMID: 31601650
- PMCID: PMC6901306
- DOI: 10.1074/jbc.RA119.010102
ERAP1 enzyme-mediated trimming and structural analyses of MHC I-bound precursor peptides yield novel insights into antigen processing and presentation
Abstract
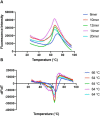
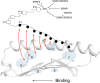
Endoplasmic reticulum aminopeptidase 1 (ERAP1) and ERAP2 critically shape the major histocompatibility complex I (MHC I) immunopeptidome. The ERAPs remove N-terminal residues from antigenic precursor peptides and generate optimal-length peptides (i.e. 8-10-mers) to fit into the MHC class I groove. It is therefore intriguing that MHC class I molecules can present N-terminally extended peptides on the cell surface that can elicit CD8+ T-cell responses. This observation likely reflects gaps in our understanding of how antigens are processed by the ERAP enzymes. To better understand ERAPs' function in antigen processing, here we generated a nested set of N-terminally extended 10-20-mer peptides (RA) n AAKKKYCL covalently bound to the human leukocyte antigen (HLA)-B*0801. We used X-ray crystallography, thermostability assessments, and an ERAP1-trimming assay to characterize these complexes. The X-ray structures determined at 1.40-1.65 Å resolutions revealed that the residue extensions (RA) n unexpectedly protrude out of the A pocket of HLA-B*0801, whereas the AAKKKYCL core of all peptides adopts similar, bound conformations. HLA-B*0801 residue 62 was critical to open the A pocket. We also show that HLA-B*0801 and antigenic precursor peptides form stable complexes. Finally, ERAP1-mediated trimming of the MHC I-bound peptides required a minimal length of 14 amino acids. We propose a mechanistic model explaining how ERAP1-mediated trimming of MHC I-bound peptides in cells can generate peptides of canonical as well as noncanonical lengths that still serve as stable MHC I ligands. Our results provide a framework to better understand how the ERAP enzymes influence the MHC I immunopeptidome.
Keywords: CD8+ T cells; HLA-B*0801; adaptive immunity; antigen presentation; antigen processing; endoplasmic reticulum (ER); endoplasmic reticulum aminopeptidase (ERAP); immunology; immunopeptidome; major histocompatibility complex I (MHC I); structural biology.
© 2019 Li et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



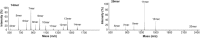

References
-
- Escobar H., Crockett D. K., Reyes-Vargas E., Baena A., Rockwood A. L., Jensen P. E., and Delgado J. C. (2008) Large scale mass spectrometric profiling of peptides eluted from HLA molecules reveals N-terminal-extended peptide motifs. J. Immunol. 181, 4874–4882 10.4049/jimmunol.181.7.4874 - DOI - PubMed
-
- Pymm P., Illing P. T., Ramarathinam S. H., O'Connor G. M., Hughes V. A., Hitchen C., Price D. A., Ho B. K., McVicar D. W., Brooks A. G., Purcell A. W., Rossjohn J., and Vivian J. P. (2017) MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat. Struct. Mol. Biol. 24, 387–394 10.1038/nsmb.3381 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

