A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function
- PMID: 31601675
- PMCID: PMC6888978
- DOI: 10.1084/jem.20190147
A novel disorder involving dyshematopoiesis, inflammation, and HLH due to aberrant CDC42 function
Abstract
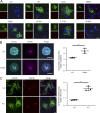
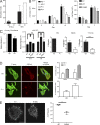
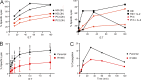
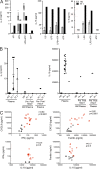
Hemophagocytic lymphohistiocytosis (HLH) is characterized by immune dysregulation due to inadequate restraint of overactivated immune cells and is associated with a variable clinical spectrum having overlap with more common pathophysiologies. HLH is difficult to diagnose and can be part of inflammatory syndromes. Here, we identify a novel hematological/autoinflammatory condition (NOCARH syndrome) in four unrelated patients with superimposable features, including neonatal-onset cytopenia with dyshematopoiesis, autoinflammation, rash, and HLH. Patients shared the same de novo CDC42 mutation (Chr1:22417990C>T, p.R186C) and altered hematopoietic compartment, immune dysregulation, and inflammation. CDC42 mutations had been associated with syndromic neurodevelopmental disorders. In vitro and in vivo assays documented unique effects of p.R186C on CDC42 localization and function, correlating with the distinctiveness of the trait. Emapalumab was critical to the survival of one patient, who underwent successful bone marrow transplantation. Early recognition of the disorder and establishment of treatment followed by bone marrow transplant are important to survival.
© 2019 Lam et al.
Figures





References
-
- Bauer C.K., Calligari P., Radio F.C., Caputo V., Dentici M.L., Falah N., High F., Pantaleoni F., Barresi S., Ciolfi A., et al. . 2018. Mutations in KCNK4 that Affect Gating Cause a Recognizable Neurodevelopmental Syndrome. Am. J. Hum. Genet. 103:621–630. 10.1016/j.ajhg.2018.09.001 - DOI - PMC - PubMed
-
- Bracaglia C., de Graaf K., Pires Marafon D., Guilhot F., Ferlin W., Prencipe G., Caiello I., Davì S., Schulert G., Ravelli A., et al. . 2017. Elevated circulating levels of interferon-γ and interferon-γ-induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 76:166–172. 10.1136/annrheumdis-2015-209020 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

