Thermodynamic control of -1 programmed ribosomal frameshifting
- PMID: 31601802
- PMCID: PMC6787027
- DOI: 10.1038/s41467-019-12648-x
Thermodynamic control of -1 programmed ribosomal frameshifting
Abstract
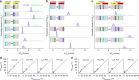
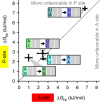
mRNA contexts containing a 'slippery' sequence and a downstream secondary structure element stall the progression of the ribosome along the mRNA and induce its movement into the -1 reading frame. In this study we build a thermodynamic model based on Bayesian statistics to explain how -1 programmed ribosome frameshifting can work. As training sets for the model, we measured frameshifting efficiencies on 64 dnaX mRNA sequence variants in vitro and also used 21 published in vivo efficiencies. With the obtained free-energy difference between mRNA-tRNA base pairs in the 0 and -1 frames, the frameshifting efficiency of a given sequence can be reproduced and predicted from the tRNA-mRNA base pairing in the two frames. Our results further explain how modifications in the tRNA anticodon modulate frameshifting and show how the ribosome tunes the strength of the base-pair interactions.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting.Nature. 2006 May 11;441(7090):244-7. doi: 10.1038/nature04735. Nature. 2006. PMID: 16688178 Free PMC article.
-
Conditional Switch between Frameshifting Regimes upon Translation of dnaX mRNA.Mol Cell. 2017 May 18;66(4):558-567.e4. doi: 10.1016/j.molcel.2017.04.023. Mol Cell. 2017. PMID: 28525745
-
A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation.Proc Natl Acad Sci U S A. 2014 Apr 15;111(15):5538-43. doi: 10.1073/pnas.1403457111. Epub 2014 Mar 31. Proc Natl Acad Sci U S A. 2014. PMID: 24706807 Free PMC article.
-
The 9-A solution: how mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting.RNA. 2003 Feb;9(2):168-74. doi: 10.1261/rna.2132503. RNA. 2003. PMID: 12554858 Free PMC article. Review.
-
P-site tRNA is a crucial initiator of ribosomal frameshifting.RNA. 2004 Feb;10(2):221-30. doi: 10.1261/rna.5122604. RNA. 2004. PMID: 14730021 Free PMC article. Review.
Cited by
-
Anticodon stem-loop tRNA modifications influence codon decoding and frame maintenance during translation.Semin Cell Dev Biol. 2024 Feb 15;154(Pt B):105-113. doi: 10.1016/j.semcdb.2023.06.003. Epub 2023 Jun 28. Semin Cell Dev Biol. 2024. PMID: 37385829 Free PMC article. Review.
-
Abracadabra, One Becomes Two: The Importance of Context in Viral -1 Programmed Ribosomal Frameshifting.mBio. 2022 Aug 30;13(4):e0246821. doi: 10.1128/mbio.02468-21. Epub 2022 Jun 23. mBio. 2022. PMID: 35735745 Free PMC article. Review.
-
Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus.Nucleic Acids Res. 2021 Nov 18;49(20):11938-11958. doi: 10.1093/nar/gkab969. Nucleic Acids Res. 2021. PMID: 34751406 Free PMC article.
-
Translational Control by Ribosome Pausing in Bacteria: How a Non-uniform Pace of Translation Affects Protein Production and Folding.Front Microbiol. 2021 Jan 11;11:619430. doi: 10.3389/fmicb.2020.619430. eCollection 2020. Front Microbiol. 2021. PMID: 33505387 Free PMC article. Review.
-
Ribosomal frameshifting selectively modulates the assembly, function, and pharmacological rescue of a misfolded CFTR variant.Proc Natl Acad Sci U S A. 2024 Oct 15;121(42):e2414768121. doi: 10.1073/pnas.2414768121. Epub 2024 Oct 10. Proc Natl Acad Sci U S A. 2024. PMID: 39388263 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

