TCRpMHCmodels: Structural modelling of TCR-pMHC class I complexes
- PMID: 31601838
- PMCID: PMC6787230
- DOI: 10.1038/s41598-019-50932-4
TCRpMHCmodels: Structural modelling of TCR-pMHC class I complexes
Abstract
The interaction between the class I major histocompatibility complex (MHC), the peptide presented by the MHC and the T-cell receptor (TCR) is a key determinant of the cellular immune response. Here, we present TCRpMHCmodels, a method for accurate structural modelling of the TCR-peptide-MHC (TCR-pMHC) complex. This TCR-pMHC modelling pipeline takes as input the amino acid sequence and generates models of the TCR-pMHC complex, with a median Cα RMSD of 2.31 Å. TCRpMHCmodels significantly outperforms TCRFlexDock, a specialised method for docking pMHC and TCR structures. TCRpMHCmodels is simple to use and the modelling pipeline takes, on average, only two minutes. Thanks to its ease of use and high modelling accuracy, we expect TCRpMHCmodels to provide insights into the underlying mechanisms of TCR and pMHC interactions and aid in the development of advanced T-cell-based immunotherapies and rational design of vaccines. The TCRpMHCmodels tool is available at http://www.cbs.dtu.dk/services/TCRpMHCmodels/ .
Conflict of interest statement
The authors declare no competing interests.
Figures

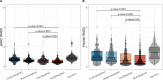
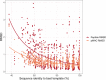





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

