Novel reassortant of H1N1 swine influenza virus detected in pig population in Russia
- PMID: 31603050
- PMCID: PMC6818105
- DOI: 10.1080/22221751.2019.1673136
Novel reassortant of H1N1 swine influenza virus detected in pig population in Russia
Abstract
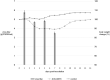
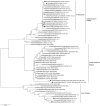
Pigs play an important role in interspecies transmission of the influenza virus, particularly as "mixing vessels" for reassortment. Two influenza A/H1N1 virus strains, A/swine/Siberia/1sw/2016 and A/swine/Siberia/4sw/2017, were isolated during a surveillance of pigs from private farms in Russia from 2016 to 2017. There was a 10% identity difference between the HA and NA nucleotide sequences of isolated strains and the most phylogenetically related sequences (human influenza viruses of 1980s). Simultaneously, genome segments encoding internal proteins were found to be phylogenetically related to the A/H1N1pdm09 influenza virus. In addition, two amino acids (129-130) were deleted in the HA of A/swine/Siberia/4sw/2017 compared to that of A/swine/Siberia/1sw/2016 HA.
Keywords: H1N1; Swine; influenza; phylogeny; reassortant.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures

References
-
- Kida H, Ito T, Yasuda J, et al. . Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75(9):2183–2188. - PubMed
-
- Krueger WS, Gray GC.. Swine influenza virus infections in man. Curr Top Microbiol Immunol. 2013;370:201–225. - PubMed
-
- Centers for Disease Control and Prevention . Information on swine influenza/variant influenza virus [Internet]. 2017 Dec 14. Available from: www.cdc.gov/flu/swineflu/index.htm
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
