Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1
- PMID: 31603676
- PMCID: PMC7737671
- DOI: 10.1021/acs.jmedchem.9b01289
Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1
Abstract
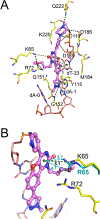
HIV-1 reverse transcriptase (RT) is an essential enzyme, targeting half of approved anti-AIDS drugs. While nucleoside RT inhibitors (NRTIs) are DNA chain terminators, the nucleotide-competing RT inhibitor (NcRTI) INDOPY-1 blocks dNTP binding to RT. Lack of structural information hindered INDOPY-1 improvement. Here we report the HIV-1 RT/DNA/INDOPY-1 crystal structure, revealing a unique mode of inhibitor binding at the polymerase active site without involving catalytic metal ions. The structure may enable new strategies for developing NcRTIs.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

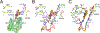
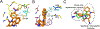
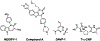
Similar articles
-
Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action.J Virol. 2006 Dec;80(24):12283-92. doi: 10.1128/JVI.00889-06. Epub 2006 Oct 4. J Virol. 2006. PMID: 17020946 Free PMC article.
-
Formation of a quaternary complex of HIV-1 reverse transcriptase with a nucleotide-competing inhibitor and its ATP enhancer.J Biol Chem. 2013 Jun 14;288(24):17336-46. doi: 10.1074/jbc.M112.433441. Epub 2013 Apr 18. J Biol Chem. 2013. PMID: 23598281 Free PMC article.
-
Impact of primer-induced conformational dynamics of HIV-1 reverse transcriptase on polymerase translocation and inhibition.J Biol Chem. 2011 Aug 26;286(34):29575-83. doi: 10.1074/jbc.M111.268235. Epub 2011 Jul 7. J Biol Chem. 2011. PMID: 21737446 Free PMC article.
-
Looking for an active conformation of the future HIV type-1 non-nucleoside reverse transcriptase inhibitors.Antivir Chem Chemother. 2010 Aug 11;20(6):213-37. doi: 10.3851/IMP1607. Antivir Chem Chemother. 2010. PMID: 20710063 Review.
-
Pyridin-2(1H)-ones: a promising class of HIV-1 non-nucleoside reverse transcriptase inhibitors.ChemMedChem. 2007 Aug;2(8):1141-7. doi: 10.1002/cmdc.200700054. ChemMedChem. 2007. PMID: 17477343 Review. No abstract available.
Cited by
-
Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.ACS Med Chem Lett. 2024 Jul 9;15(8):1232-1241. doi: 10.1021/acsmedchemlett.4c00117. eCollection 2024 Aug 8. ACS Med Chem Lett. 2024. PMID: 39140041 Free PMC article.
-
Insights into HIV-1 Reverse Transcriptase (RT) Inhibition and Drug Resistance from Thirty Years of Structural Studies.Viruses. 2022 May 11;14(5):1027. doi: 10.3390/v14051027. Viruses. 2022. PMID: 35632767 Free PMC article. Review.
-
Avoiding Drug Resistance in HIV Reverse Transcriptase.Chem Rev. 2021 Mar 24;121(6):3271-3296. doi: 10.1021/acs.chemrev.0c00967. Epub 2021 Jan 28. Chem Rev. 2021. PMID: 33507067 Free PMC article. Review.
-
Selection of Primer-Template Sequences That Bind with Enhanced Affinity to Vaccinia Virus E9 DNA Polymerase.Viruses. 2022 Feb 10;14(2):369. doi: 10.3390/v14020369. Viruses. 2022. PMID: 35215961 Free PMC article.
-
Indole alkaloids inhibit zika and chikungunya virus infection in different cell lines.BMC Complement Med Ther. 2021 Aug 28;21(1):216. doi: 10.1186/s12906-021-03386-z. BMC Complement Med Ther. 2021. PMID: 34454481 Free PMC article.
References
-
- Piot P; Abdool Karim SS; Hecht R; Legido-Quigley H; Buse K; Stover J; Resch S; Ryckman T; Mogedal S; Dybul M; Goosby E; Watts C; Kilonzo N; McManus J; Sidibe M; UNAIDS—Lancet Commission.. Defeating AIDS—advancing global health. Lancet 2015, 386, 171–218. - PubMed
-
- Waheed AA; Tachedjian G Why Do We Need New Drug Classes for HIV Treatment and Prevention? Curr. Top. Med. Chem 2016, 16, 1343–1349. - PubMed
-
- Jochmans D; Deval J; Kesteleyn B; Van Marck H; Bettens E; De Baere I; Dehertogh P; Ivens T; Van Ginderen M; Van Schoubroeck B; Ehteshami M; Wigerinck P; Gotte M; Hertogs K Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J Virol 2006, 80, 12283–12292. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

