Deep Neural Networks Improve Radiologists' Performance in Breast Cancer Screening
- PMID: 31603772
- PMCID: PMC7427471
- DOI: 10.1109/TMI.2019.2945514
Deep Neural Networks Improve Radiologists' Performance in Breast Cancer Screening
Abstract
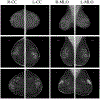
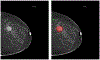
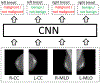
We present a deep convolutional neural network for breast cancer screening exam classification, trained, and evaluated on over 200000 exams (over 1000000 images). Our network achieves an AUC of 0.895 in predicting the presence of cancer in the breast, when tested on the screening population. We attribute the high accuracy to a few technical advances. 1) Our network's novel two-stage architecture and training procedure, which allows us to use a high-capacity patch-level network to learn from pixel-level labels alongside a network learning from macroscopic breast-level labels. 2) A custom ResNet-based network used as a building block of our model, whose balance of depth and width is optimized for high-resolution medical images. 3) Pretraining the network on screening BI-RADS classification, a related task with more noisy labels. 4) Combining multiple input views in an optimal way among a number of possible choices. To validate our model, we conducted a reader study with 14 readers, each reading 720 screening mammogram exams, and show that our model is as accurate as experienced radiologists when presented with the same data. We also show that a hybrid model, averaging the probability of malignancy predicted by a radiologist with a prediction of our neural network, is more accurate than either of the two separately. To further understand our results, we conduct a thorough analysis of our network's performance on different subpopulations of the screening population, the model's design, training procedure, errors, and properties of its internal representations. Our best models are publicly available at https://github.com/nyukat/breast_cancer_classifier.
Figures





References
-
- Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2015,” CA, Cancer J. Clinicians, vol. 65, no. 1, pp. 5–29, 2015. - PubMed
-
- Duffy SW et al. , “The impact of organized mammography service screening on breast carcinoma mortality in seven Swedish counties: A collaborative evaluation,” Cancer, vol. 95, no. 3, pp. 458–469, 2002. - PubMed
-
- Kopans DB, “Beyond randomized controlled trials: Organized mammographic screening substantially reduces breast carcinoma mortality,” Cancer, vol. 94, no. 2, pp. 580–581, 2002. - PubMed
-
- Duffy SW, Tabár L, and Smith RA, “The mammographic screening trials: Commentary on the recent work by Olsen and Gøtzsche,” CA, Cancer J. Clinicians, vol. 52, no. 2, pp. 68–71, 2002. - PubMed
-
- Kopans DB, “An open letter to panels that are deciding guidelines for breast cancer screening,” Breast Cancer Res Treat, vol. 151, no. 1, pp. 19–25, 2015. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

